参数修改
任务切换
全局修改
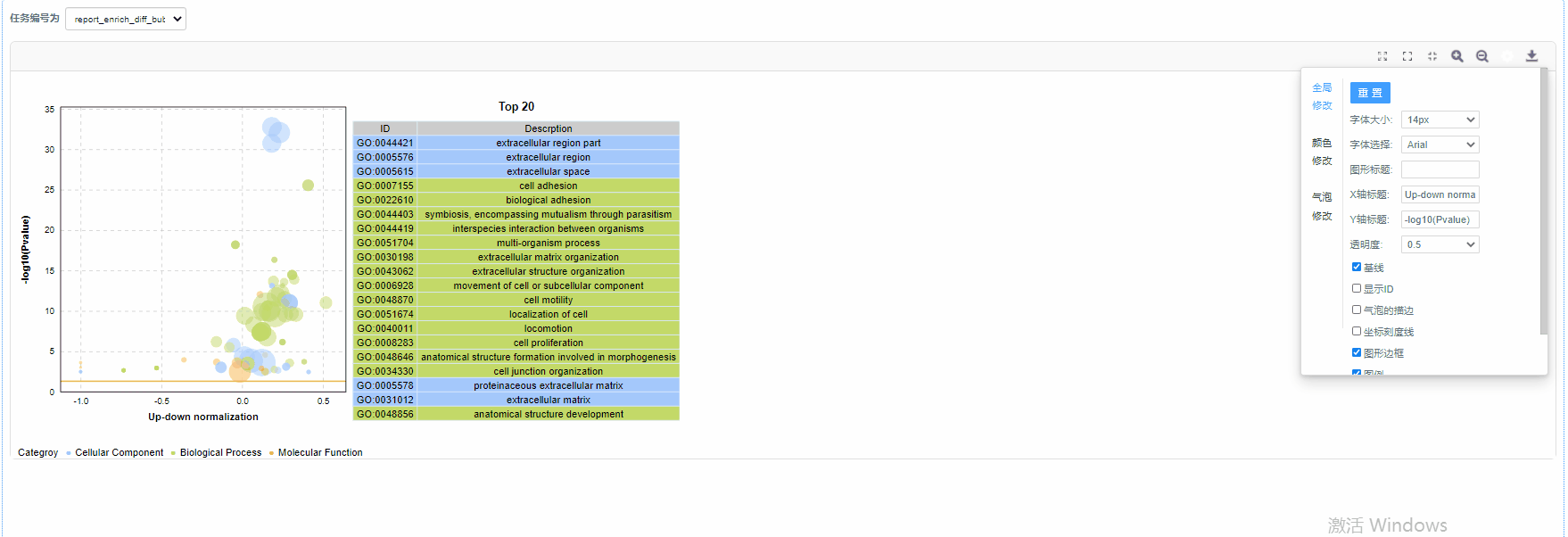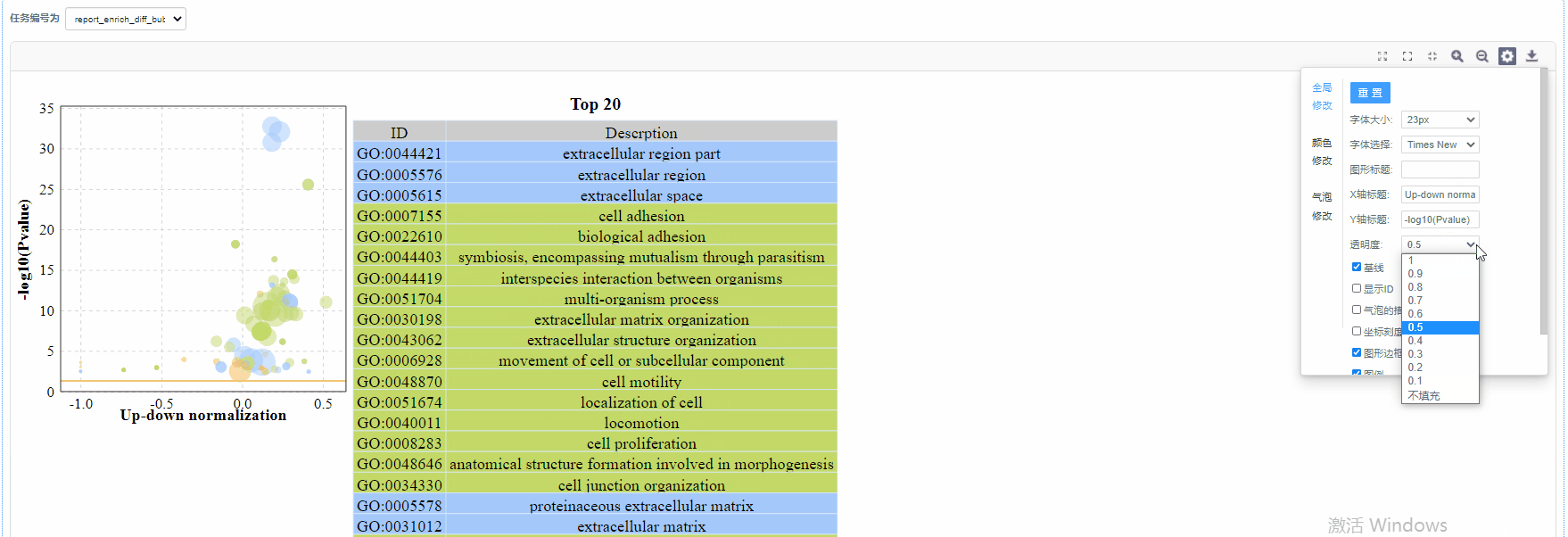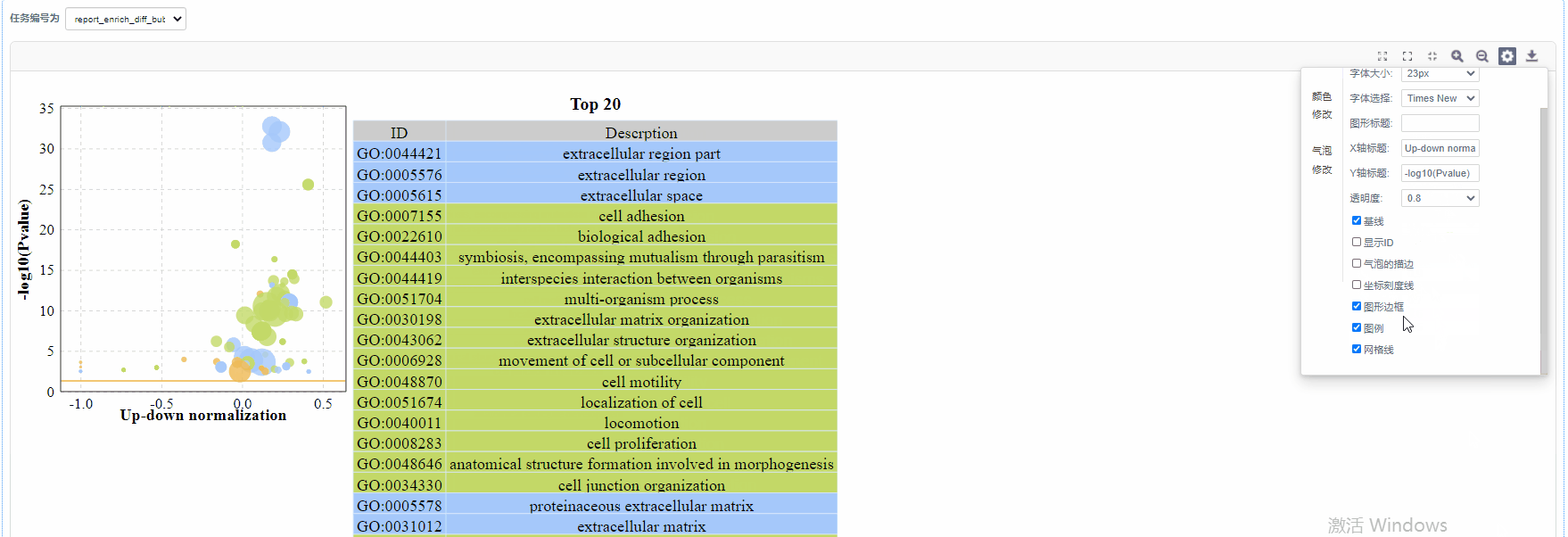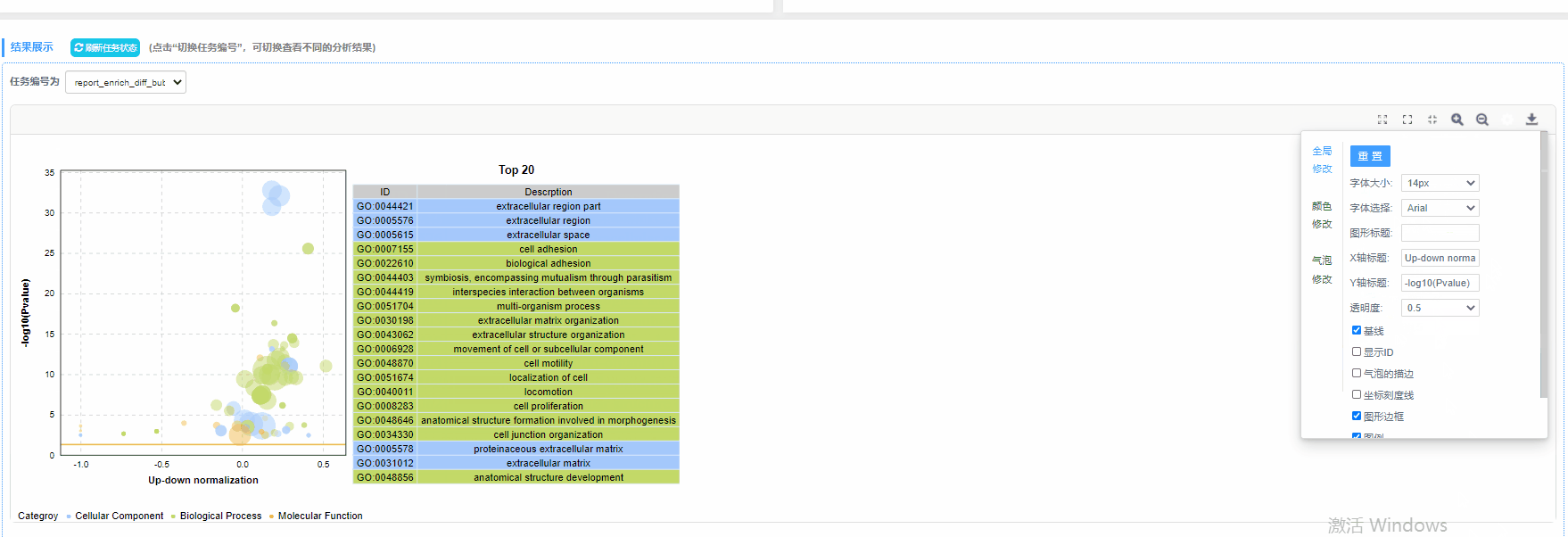
颜色修改
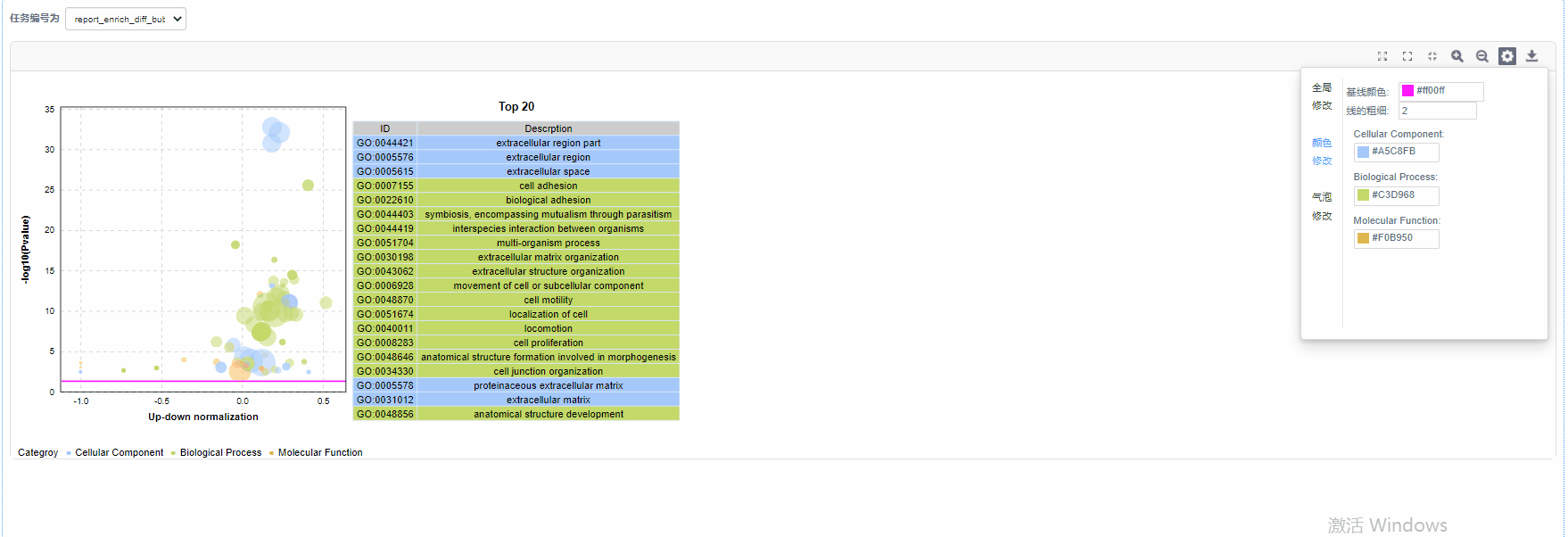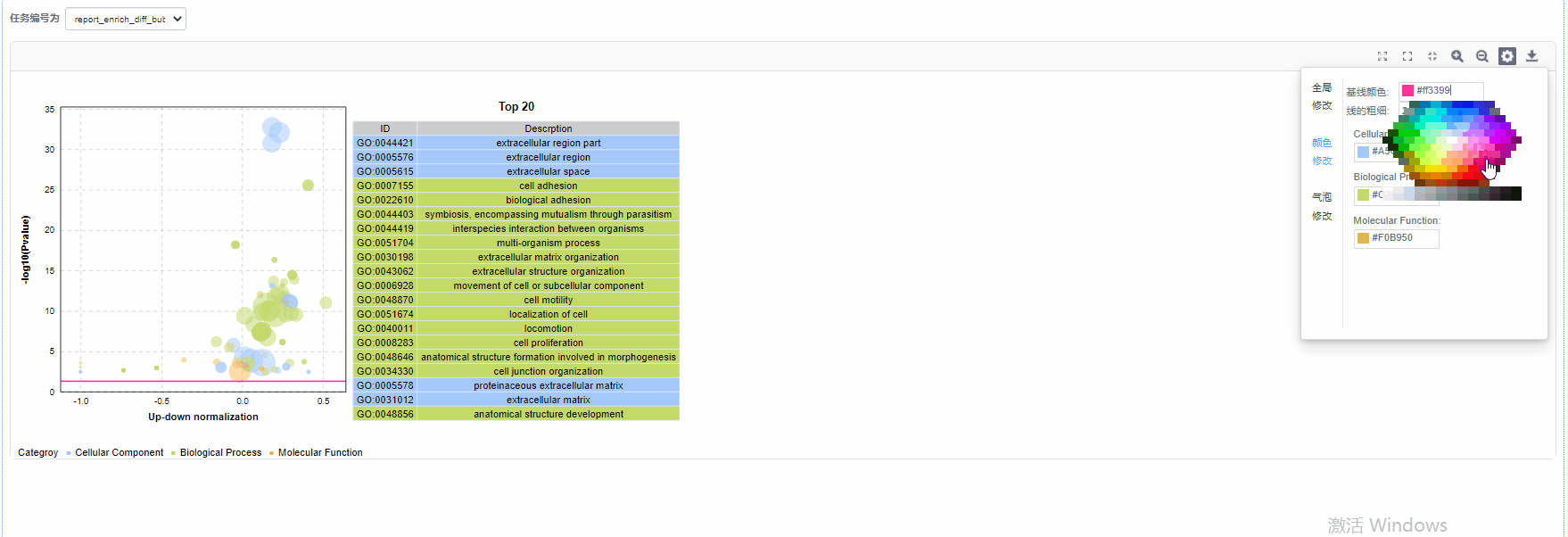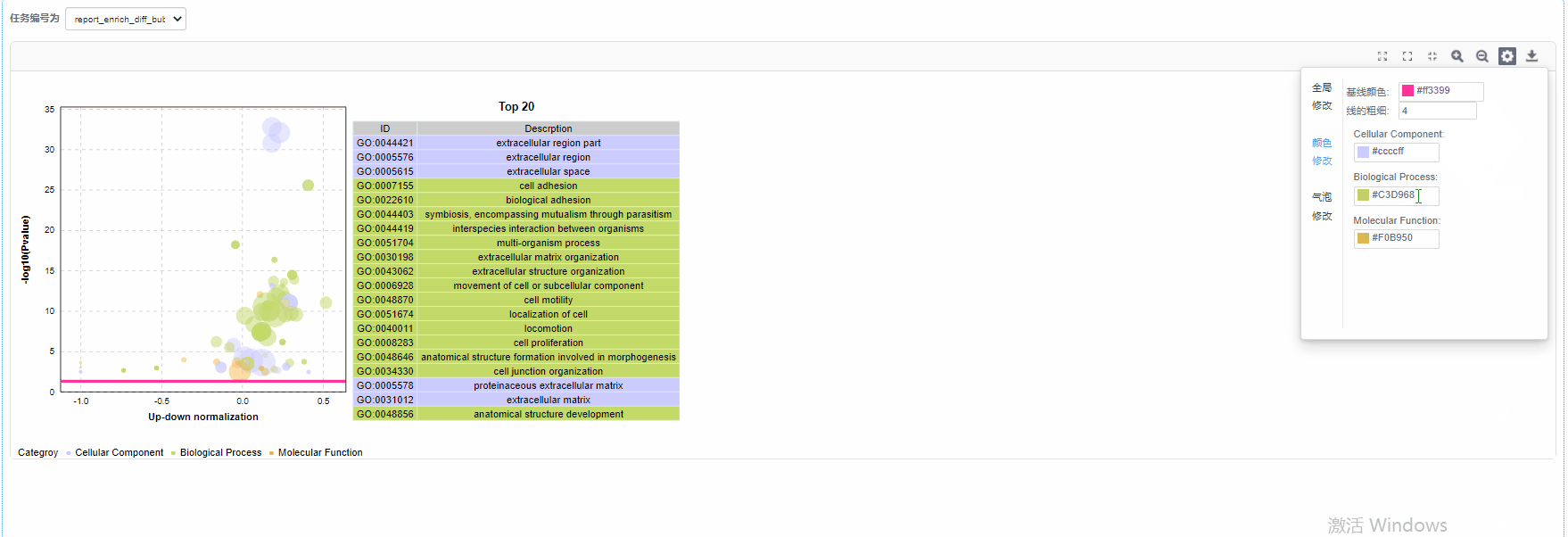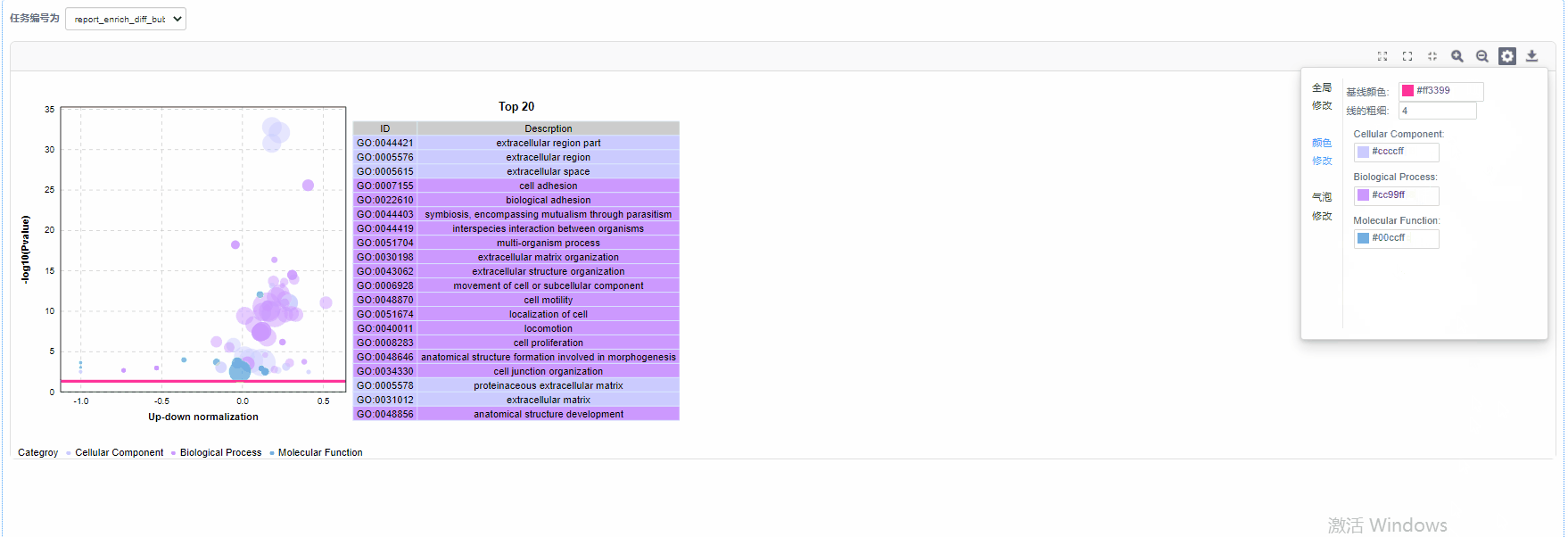
气泡修改
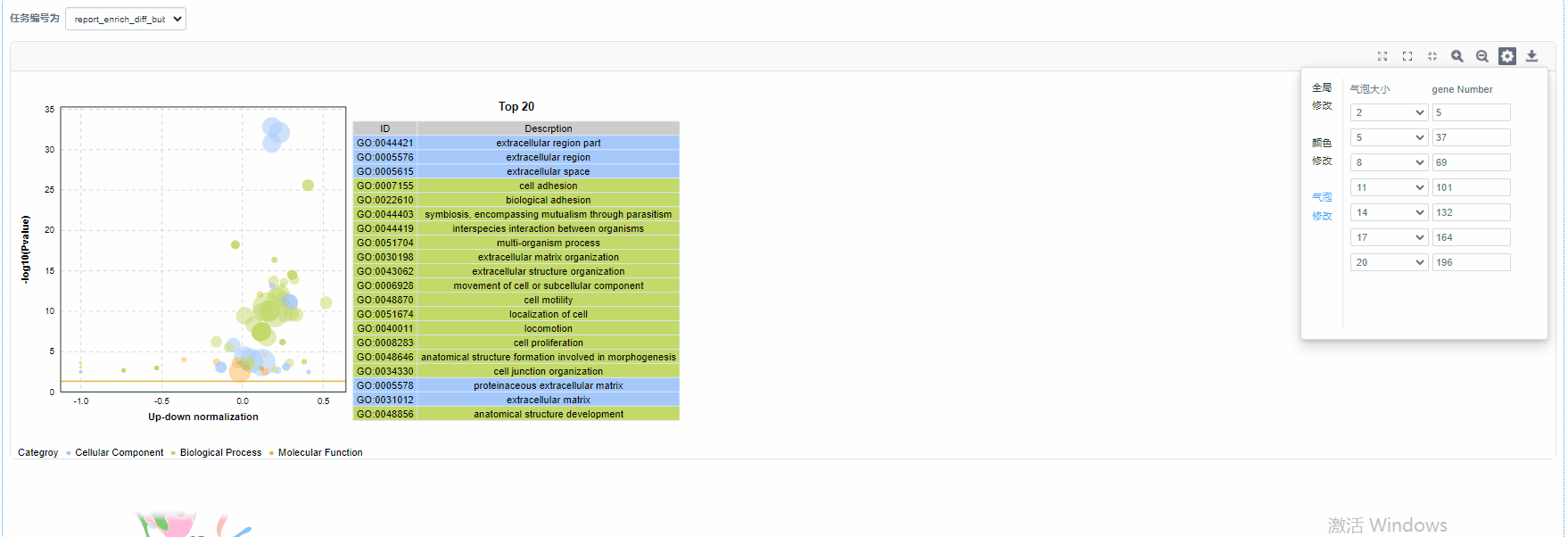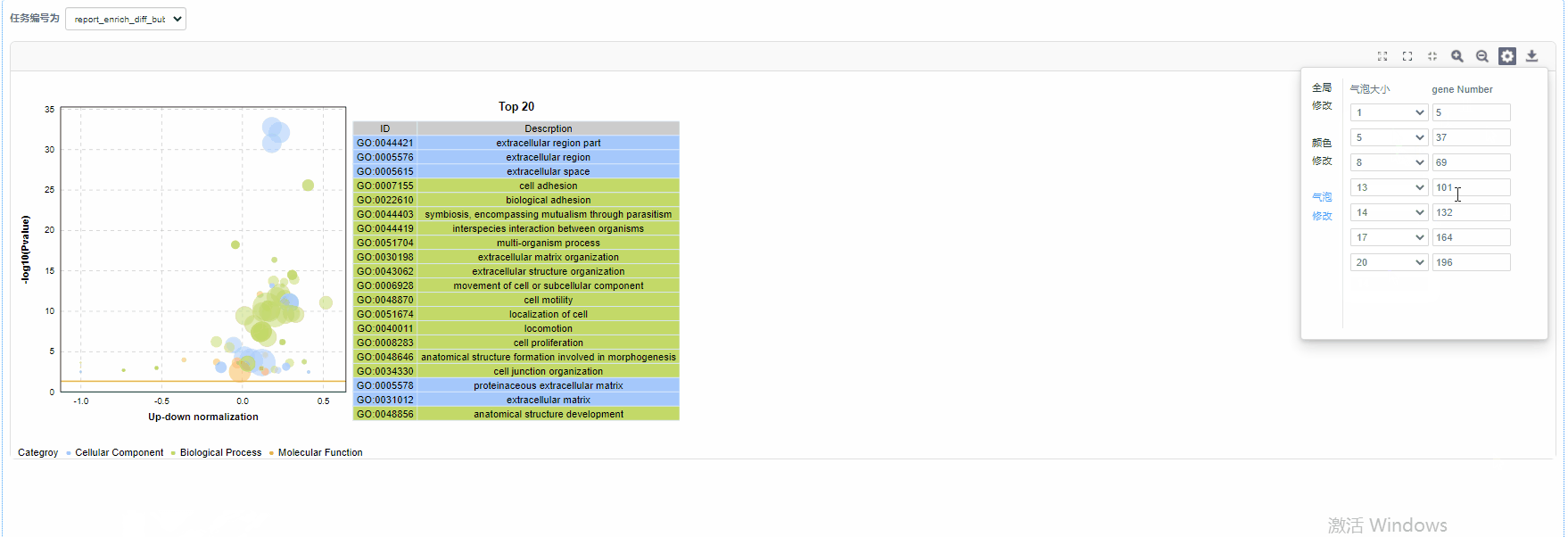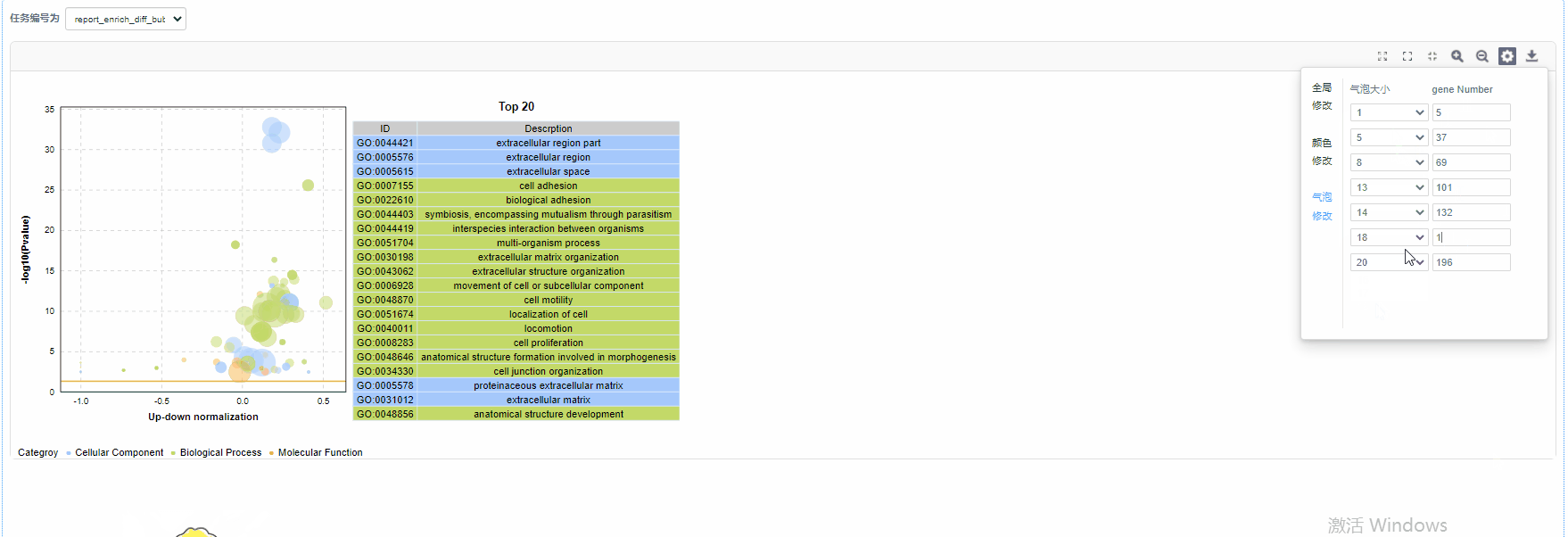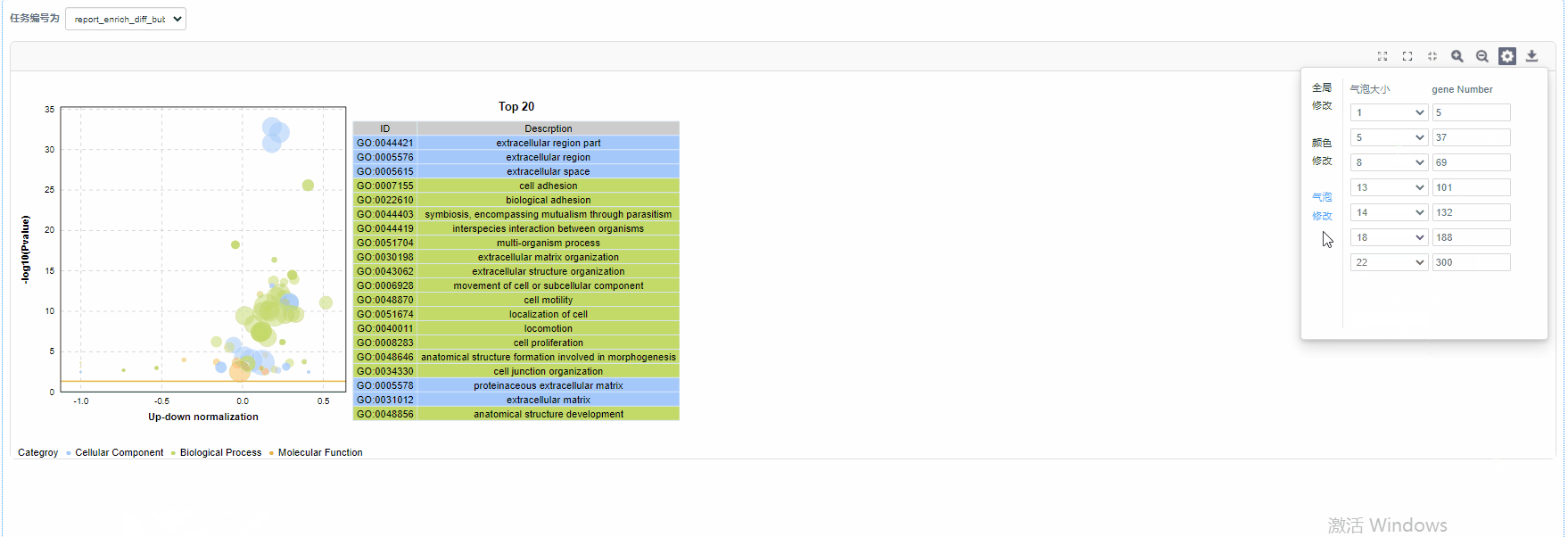
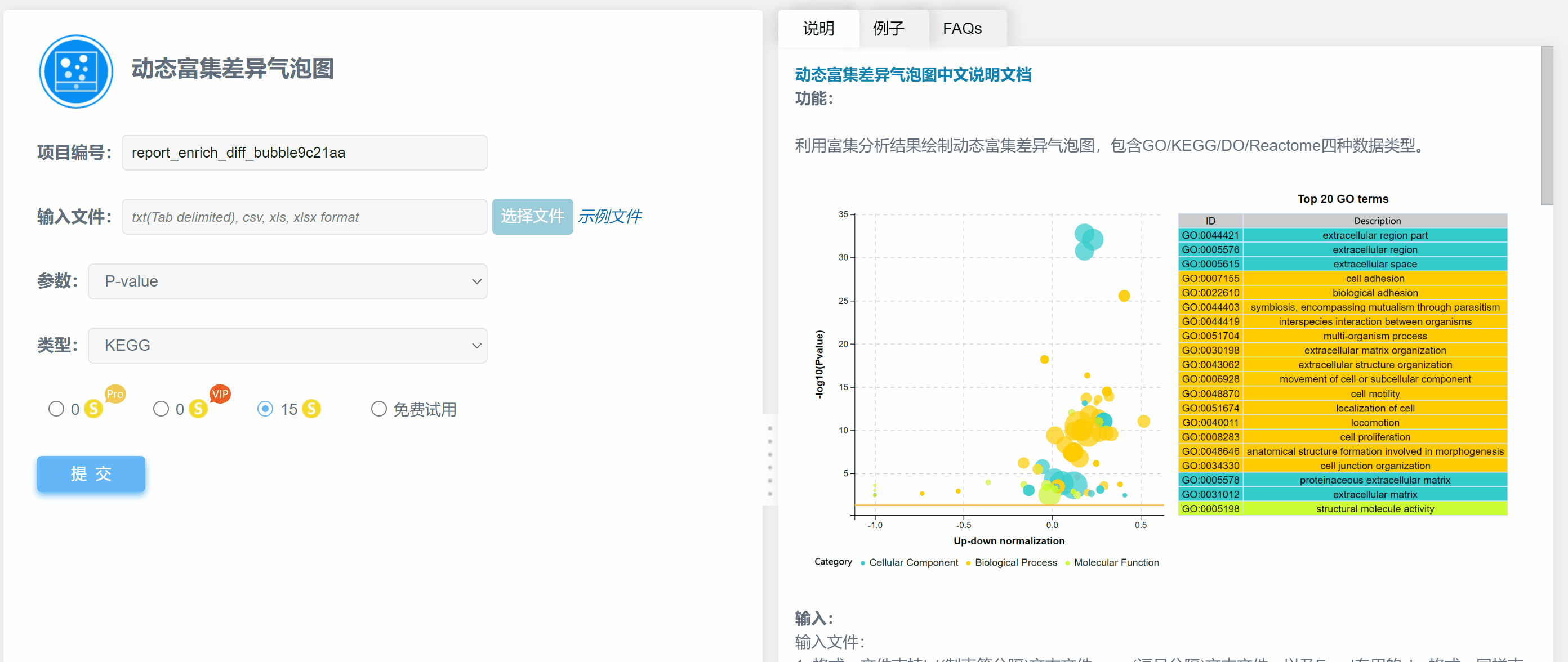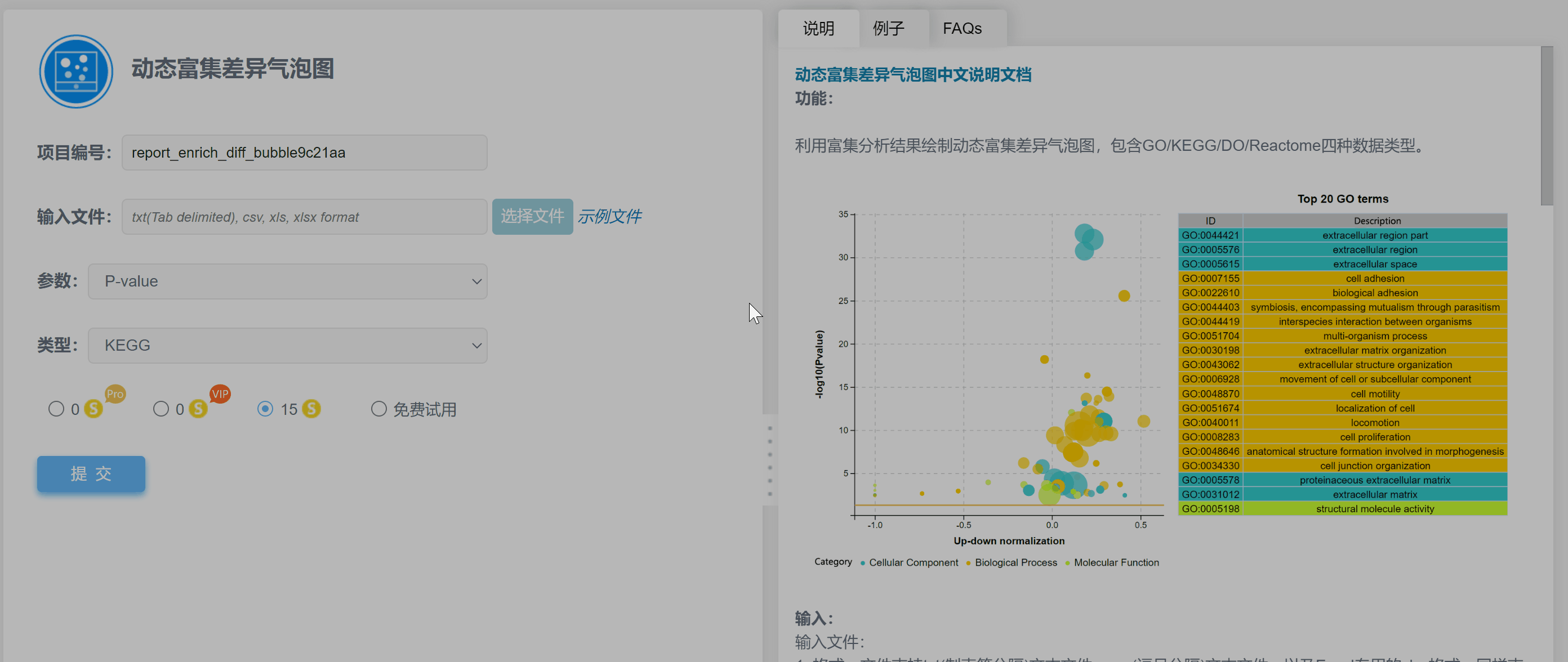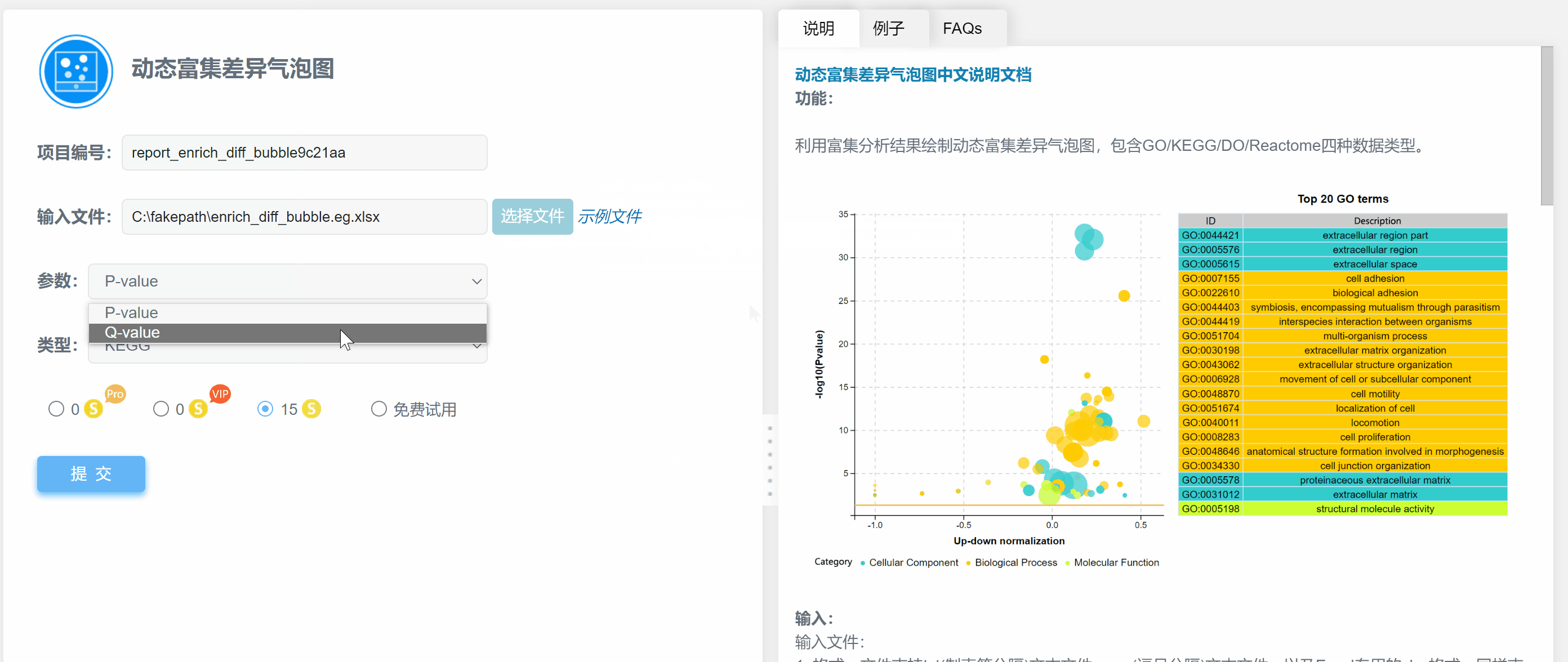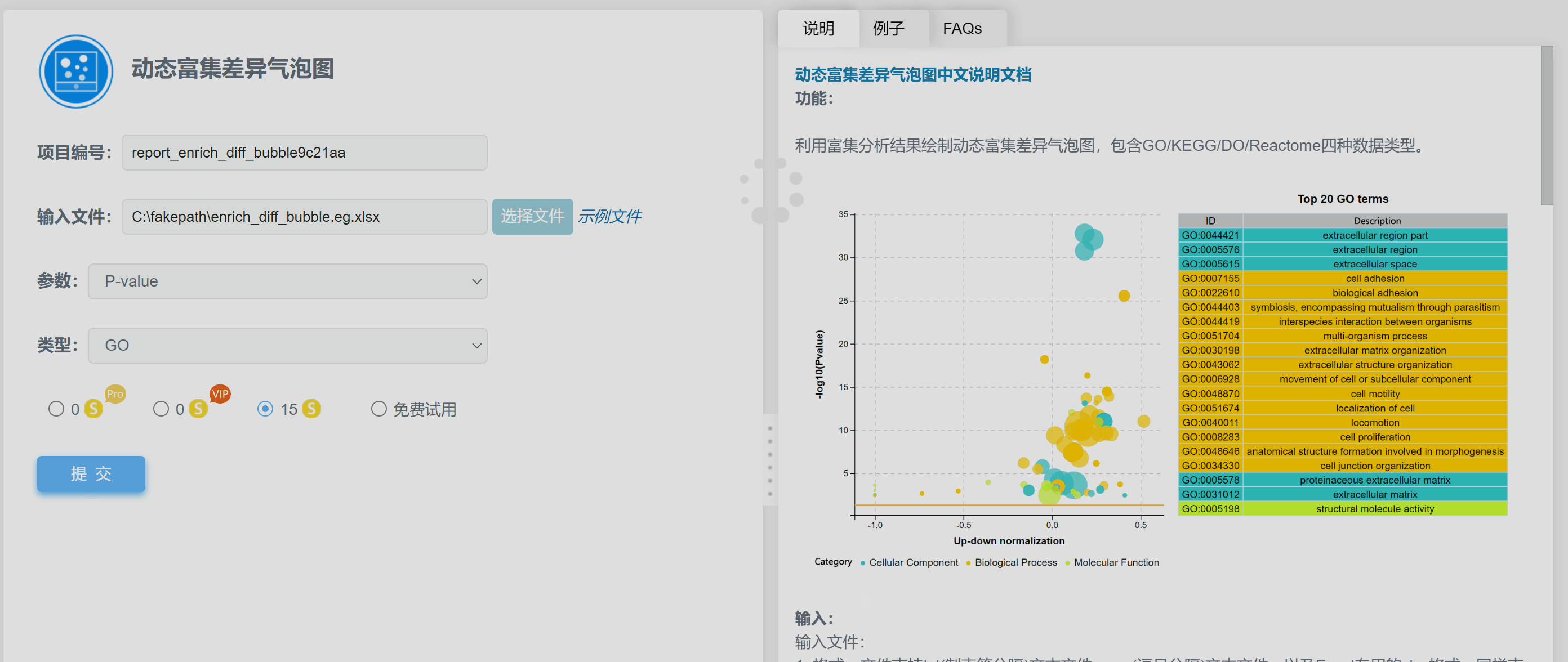
功能:
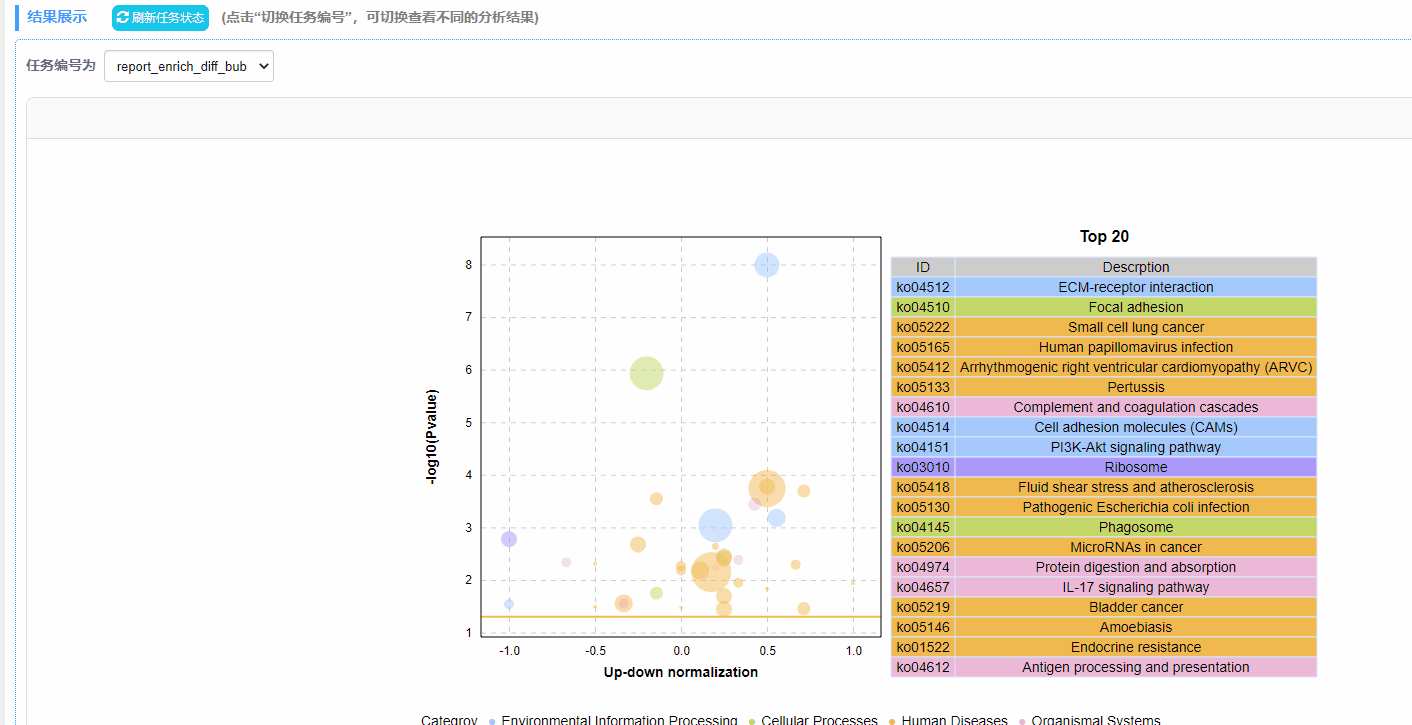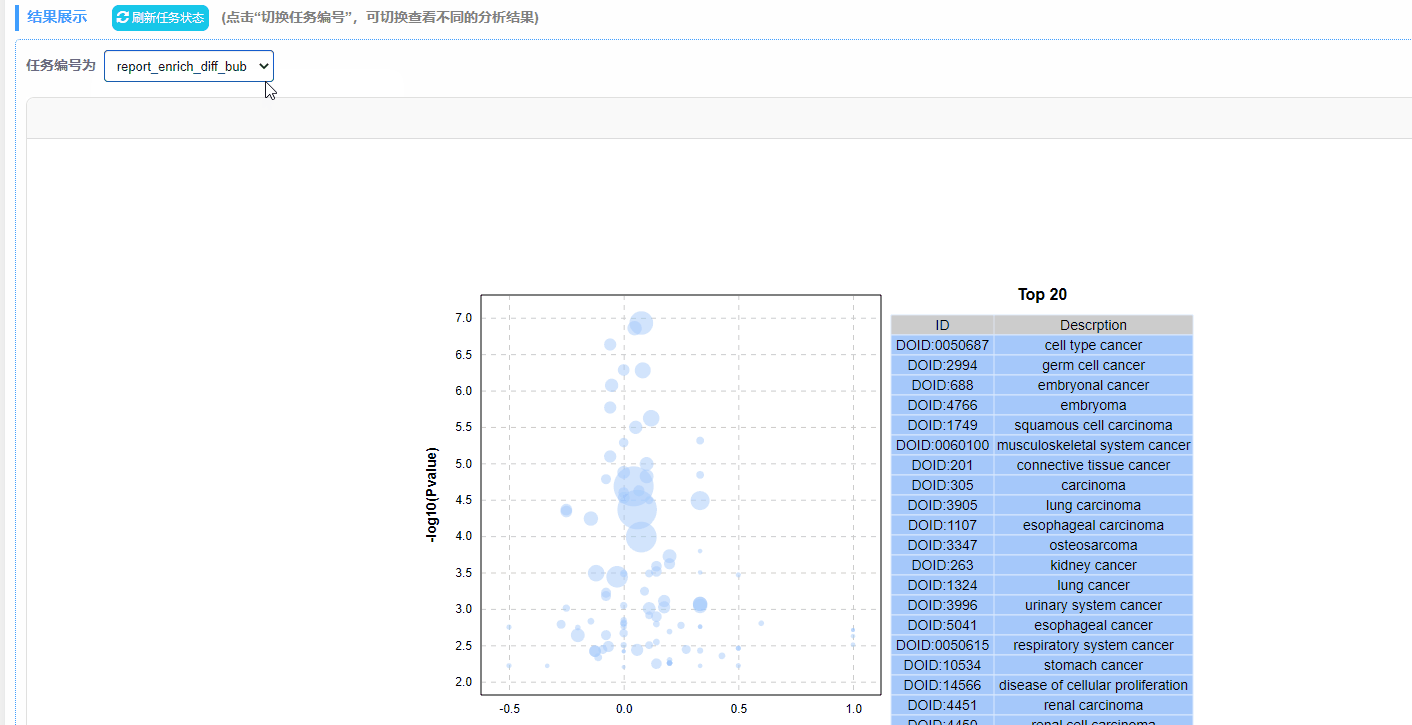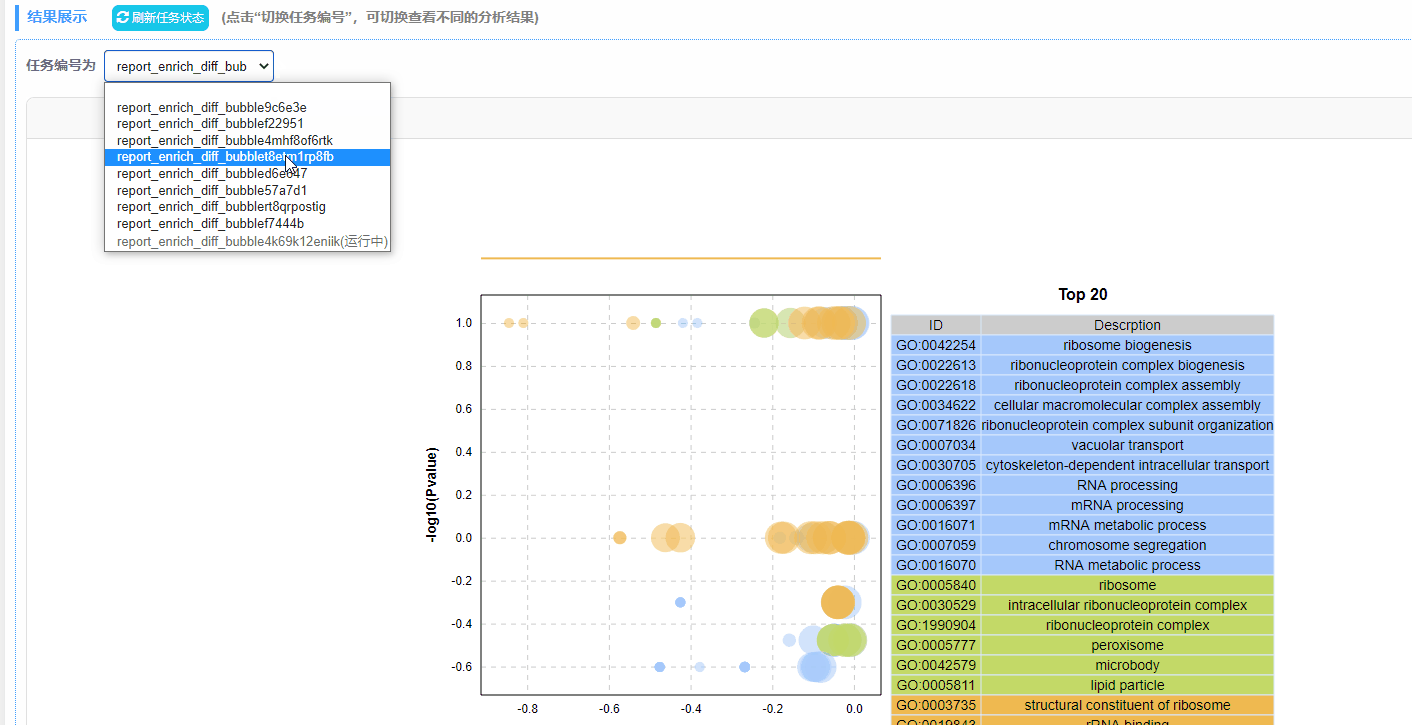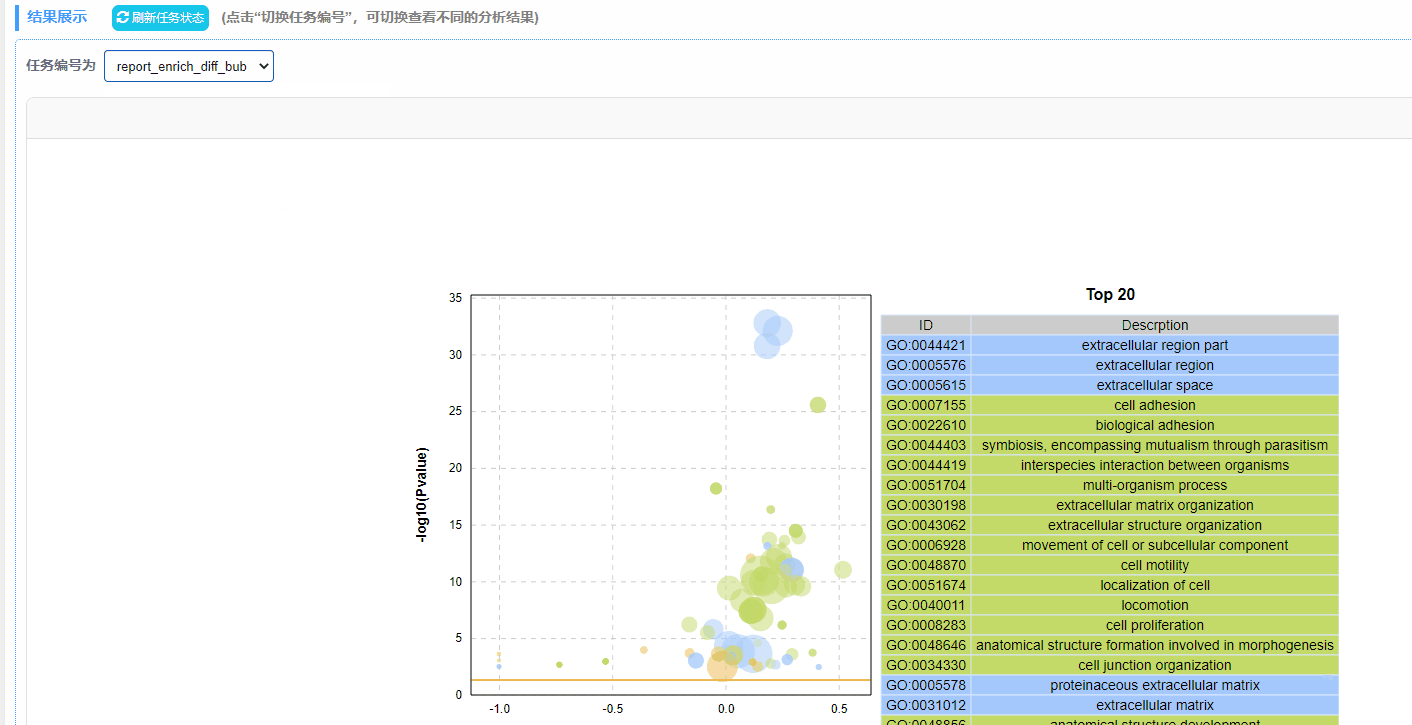
利用富集分析结果绘制动态富集差异气泡图,包含GO/KEGG/DO/Reactome四种数据类型。
输入:
输入文件:
1. 格式:文件支持txt(制表符分隔)文本文件、csv(逗号分隔)文本文件、以及Excel专用的xlsx格式,同样支持旧版Excel的xls(Excel 97-2003 )格式。
文件名可由英文和数字构成,文件拓展名没有限制,可以是“.txt”、“.xlsx”、“.xls”、“.csv” 等,例如 mydata01.txt,gene02.xlsx 。
2. 绘图方式:以列号形式读取数据,规定第4列为差异上调基因数目,第5列为差异下调基因数目,第6列为P/Q值,筛选P/Q值最小的前20行数据绘制气泡图。
3. 文件类型:
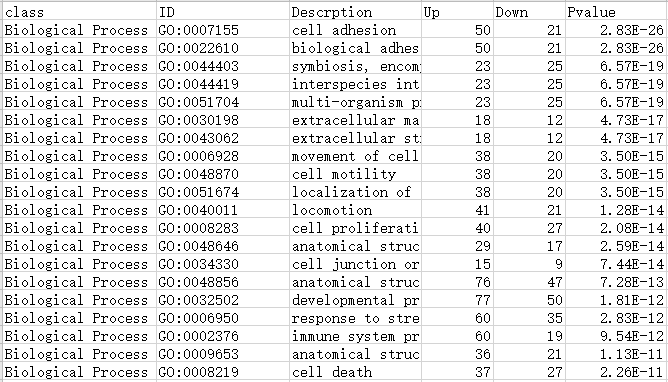
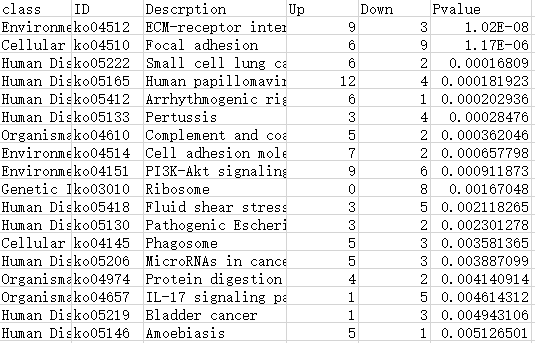
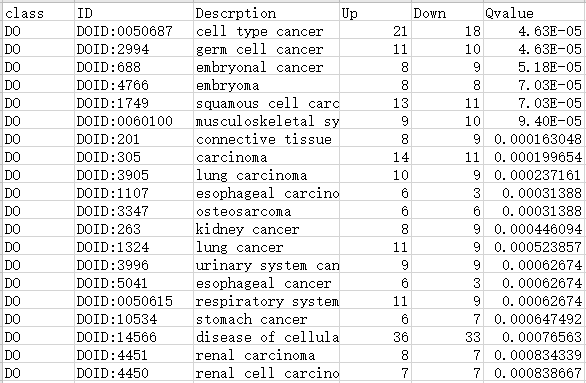
Type1: 包含6列数据
第1列:一级/A级分类
第2列:GO/KEGG/DO/Reactome注释ID号
第3列:Term/Pathway名称
第4列:属于这个通路的差异上调基因数
第5列:属于这个通路的差异下调基因数
第6列:富集分析的P值/Q值
注:最终绘制的图中,横坐标up-down normalization=(差异上调基因数-差异下调基因数)/(差异上调基因数+差异下调基因数)
表格如下表所示:
GO数据库
KEGG数据库
DO数据库
Reactome数据库
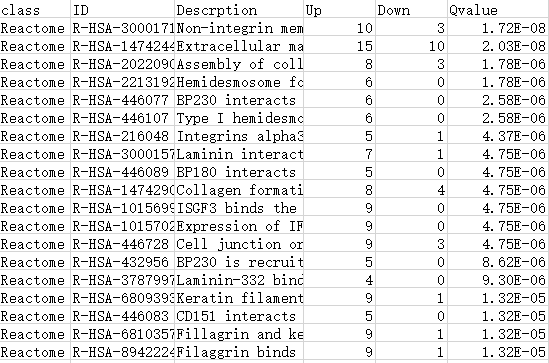
Type2: 包含更多数据信息
第1列:一级/A级分类
第2列:GO/KEGG/DO/Reactome注释ID号
第3列:Term/Pathway名称
第4列:属于这个通路的差异上调基因数
第5列:属于这个通路的差异下调基因数
第6列:富集分析的P值/Q值
第7列:差异上调基因ID
第8列:差异下调基因ID
注:最终绘制的图中,横坐标up-down normalization=(差异上调基因数-差异下调基因数)/(差异上调基因数+差异下调基因数)
表格如下表所示:
1. 参数选择:
1)参数:根据需求选择使用P-value或Q-value绘制气泡图,选择不同的参数将会影响最后的图形结果;
2)类型:包含GO、KO、DO、Reactome四大富集分析数据库,根据表格数据来源选择对应类型。
2. 参数调整:
1)图形标题:设置或修改图形的标题
2)X/Y轴标题:设置或修改X/Y轴标题
3)透明度:设置气泡的透明度
4)显示ID/气泡的描边/坐标刻度线/图形边框/图例/网格线:设置是否展示
5)基线修改:自定义设置基线颜色以及基线粗细
6)气泡颜色修改:设置GO/KO/DO/Reactome一级分类颜色
7)气泡大小修改:设置图形中气泡的大小
8)重置:恢复图片的默认设置
3. 输出:
程序将按要求输出富集差异气泡图,用户可根据个人需求进行参数调整,可提供SVG和PNG两种格式的结果文件下载。
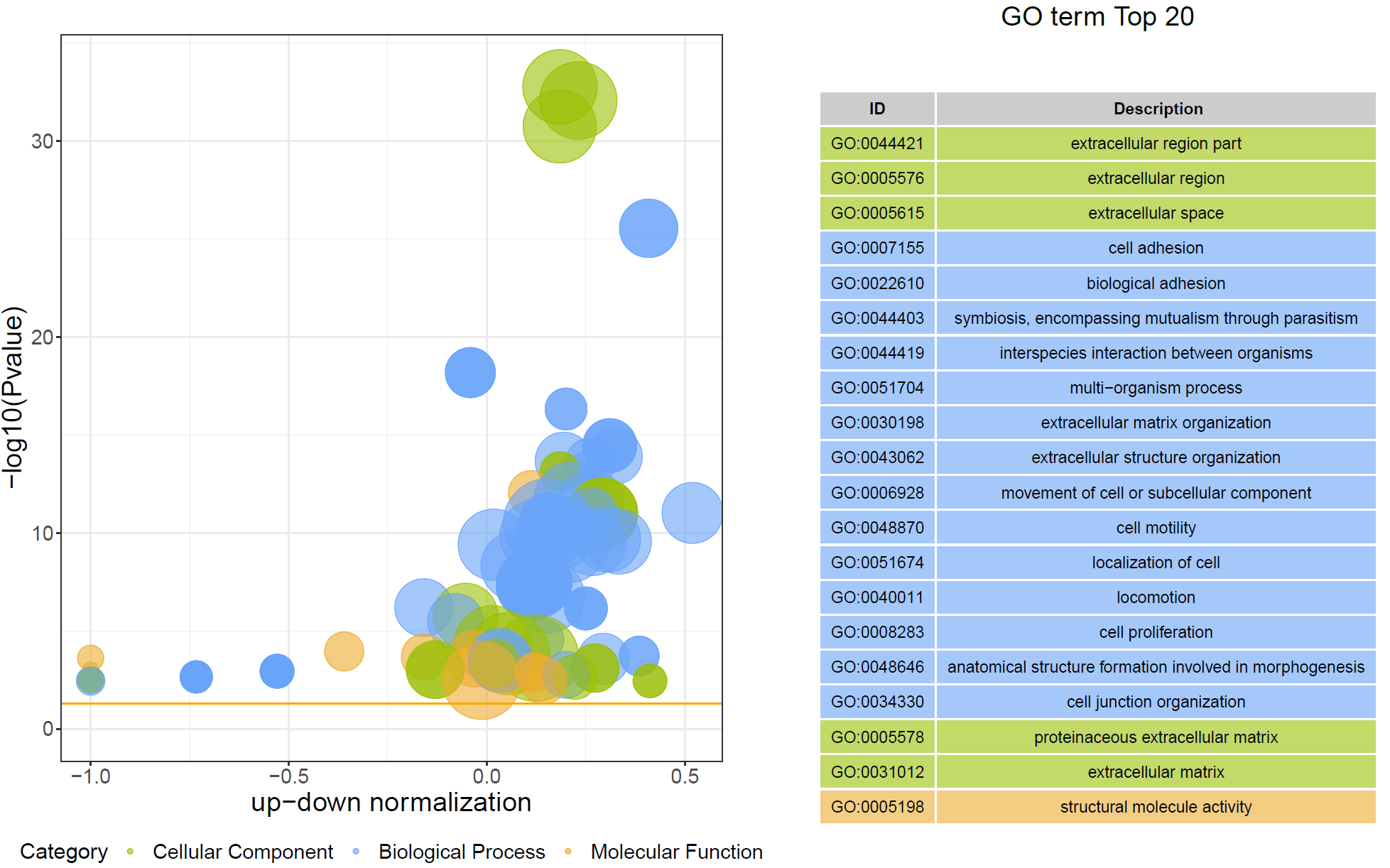
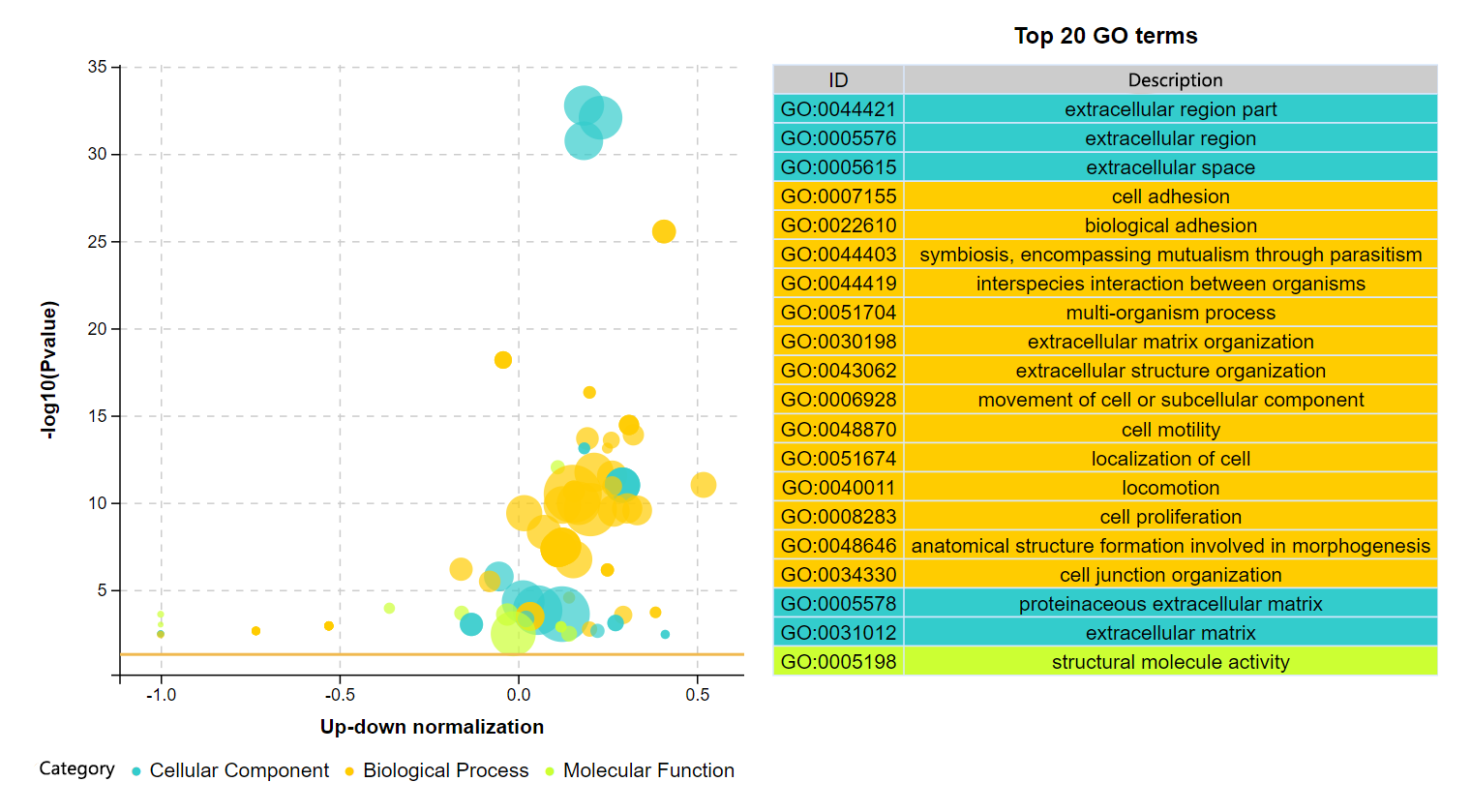
结果说明:
纵坐标为-log10(Q/Pvalue),横坐标为up-down normalization值(上调基因数目与下调基因数目的差值占总差异基因的比例);气泡大小表示当前term(pathway)富集到的目的基因数;黄线代表Q/Pvalue=0.05的阈值;右边为Q/P值前20的term/Pathway列表,不同的颜色代表不同的Ontology/A class。
Q1. 上传的数据需要保存成什么格式?文件名称和拓展名有没有要求?
OmicShare当前支持txt(制表符分隔)文本文件、csv(逗号分隔)文本文件、以及Excel专用的xlsx格式,同样支持旧版Excel的xls(Excel 97-2003 )格式。如果是核酸、蛋白序列文件,必须为FASTA格式(本质是文本文件)。
文件名可由英文和数字构成,文件拓展名没有限制,可以是“.txt”、“.xlsx”、“.xls”、“.csv”“.fasta”等,例如 mydata01.txt,gene02.xlsx 。
Q2. 提交时报错常见问题:
1.提交时显示X行X列空行/无数据,请先自查表格中是否存在空格或空行,需要删掉。
2.提交时显示列数只有1列,但表格数据不止1列:列间需要用分隔符隔开,先行检查文件是否用了分隔符。
其它提示报错,请先自行根据提示修改;如果仍然无法提交,可通过左侧导航栏的“联系客服”选项咨询OmicShare客服。
Q3. 提交的任务完成后却不出图该怎么办?
主要原因是上传的数据文件存在特殊符号所致。可参考以下建议逐一排查出错原因:
(1)数据中含中文字符,把中文改成英文;
(2) 数据中含特殊符号,例如 %、NA、+、-、()、空格、科学计数、罗马字母等,去掉特殊符号,将空值用数字“0”替换;
(3)检查数据中是否有空列、空行、重复的行、重复的列,特别是行名(一般为gene id)、列名(一般为样本名)出现重复值,如果有删掉。
排查完之后,重新上传数据、提交任务。如果仍然不出图,可通过左侧导航栏的“联系客服”选项咨询OmicShare客服。
Q4.下载的结果文件用什么软件打开?
OmicShare云平台的结果文件(例如,下图为KEGG富集分析的结果文件)包括两种类型:图片文件和文本文件。
图片文件:
为了便于用户对图片进行后期编辑,OmicShare同时提供位图(png)和矢量图(pdf、svg)两种类型的图片。对于矢量图,最常见的是pdf和svg格式,常用Ai(Adobe illustrator)等进行编辑。其中,svg格式的图片可用网页浏览器打开,也可直接在word、ppt中使用。
文本文件:
文本文件的拓展名主要有4种类型:“.os”、“.xls”、“.log”和“.txt”。这些文件本质上都是制表符分隔的文本文件,使用记事本、Notepad++、EditPlus、Excel等文本编辑器直接打开即可。结果文件中,拓展名为“.os”文件为上传的原始数据;“.xls”文件一般为分析生成的数据表格;“.log”文件为任务运行日志文件,便于检查任务出错原因。
Q5. 提交的任务一直在排队怎么办?
提交任务后都需要排队,1分钟后,点击“任务状态刷新”按钮即可。除了可能需运行数天的注释工具,一般工具数十秒即可出结果,如果超出30分钟仍无结果,请联系OS客服,发送任务编号给OmicShare客服,会有专人为你处理任务问题。
Q6. 结果页面窗口有问题,图表加载不出来怎么办?
尝试用谷歌浏览器登录OmicShare查看结果文件,部分浏览器可能不兼容。
引用OmicShare Tools的参考文献为:
Mu, Hongyan, Jianzhou Chen, Wenjie Huang, Gui Huang, Meiying Deng, Shimiao Hong, Peng Ai, Chuan Gao, and Huangkai Zhou. 2024. “OmicShare tools: a Zero‐Code Interactive Online Platform for Biological Data Analysis and Visualization.” iMeta e228. https://doi.org/10.1002/imt2.228案例1:
发表期刊:Animals
影响因子:3.0
发表时间:2022
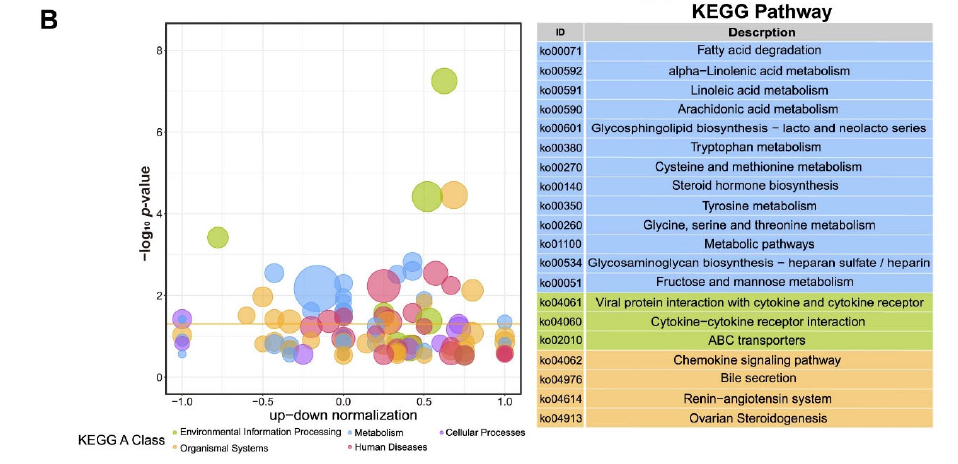
Figure 3. GO and KEGG analysis of DEGs for Duroc and Luchuan adipose. (B) KEGG summary graph showing the summary of the KEGG pathway. Different colors represent different KEGG A class categories.
引用方式:
Gene expression level was measured by Fragments per Kilobase per Million Mapped Fragments (FPKM) [17], the most commonly used method for estimating gene expression abundance. DEGSeq [18] was used to analyze DEGs (|Fold Change| > 2 and p-value < 0.01).The Principal Component Analysis (PCA), Gene Ontology (GO) term enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and GSEA were performed using Omicshare, a real-time interactive online data analysis platform (p-value < 0.05) (http://www.omicshare.com (accessed on 18 February 2022)) [19].
参考文献:
Pan H, Huang T, Yu L, et al. Transcriptome Analysis of the Adipose Tissue of Luchuan and Duroc Pigs[J]. Animals, 2022, 12(17): 2258.
案例2:
发表期刊:Frontiers in Veterinary Science
影响因子:3.2
发表时间:2022
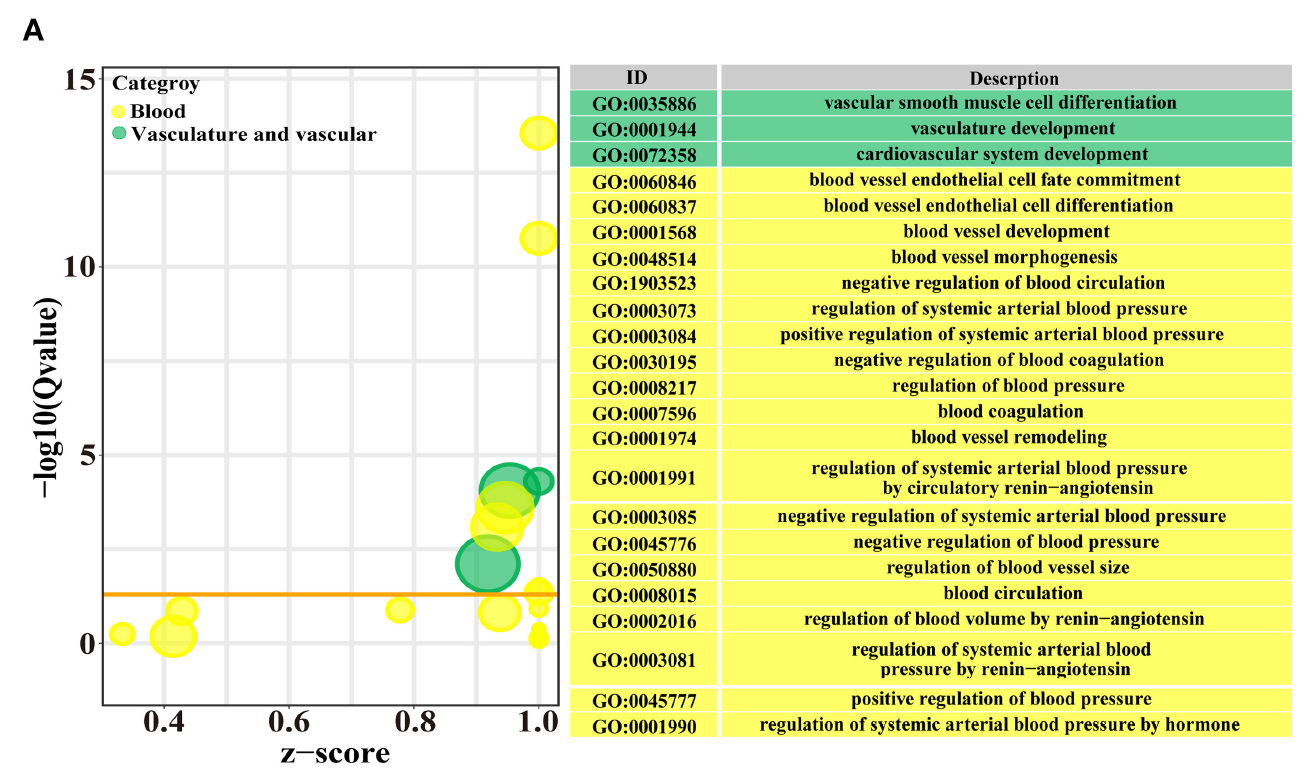
FIGURE 3. The candidate DEPs selected from the 3,739 DEPs and GO terms related to angiogenesis. (A) the GO terms selected from the 819 significantly different biological processes related to blood, vascular or vasculature, and angiogenesis in the DIA data.
引用方式:
The heat maps, Volcano plots, bubble and Upset Venn diagrams were drawn using the R language and the online OmicShare tools (https://www.omicshare.com/tools/) (17, 18).
参考文献:
Zhang Q, Bai X, Shi J, et al. DIA proteomics identified the potential targets associated with angiogenesis in the mammary glands of dairy cows with hemorrhagic mastitis[J]. Frontiers in Veterinary Science, 2022, 9: 980963.




 扫码支付更轻松
扫码支付更轻松



