适用范围:
可对18个常见物种的基因集进行富集分析,牛、斑马鱼、人、猕猴、小鼠、大鼠、猪、秀丽线虫、果蝇、拟南芥、水稻、番茄、小麦、玉米、酵母、山羊、鸡、籼稻,并且提供3个基因组版本;
也可以自行准备研究物种的背景基因进行富集分析。
输入:
①输入的表格文件支持txt(制表符分隔)文本文件、csv(逗号分隔)文本文件、以及Excel专用的xlsx格式,同样支持旧版Excel的xls(Excel 97-2003 )格式。也可以输入两列,diff是差异结果,第二列为差异倍数取log2,大于0上调,小于0下调,此时参数应选择diff。
②富集的目的基因列表,即想要研究的基因列表,每行第一列为基因id,基因id要包含在背景基因表中。
③背景基因总表,如果是有参考基因组的模式生物,可以直接使用已有参考基因作为背景基因文件。目前提供的物种有水稻、拟南芥、小鼠、大鼠、斑马鱼、鸡、秀丽线虫、果蝇、人。ID类型可选择基因ID或转录本ID,根据富集目的基因的ID类型决定。可以点击“预览参考文件”来查看具体ID。
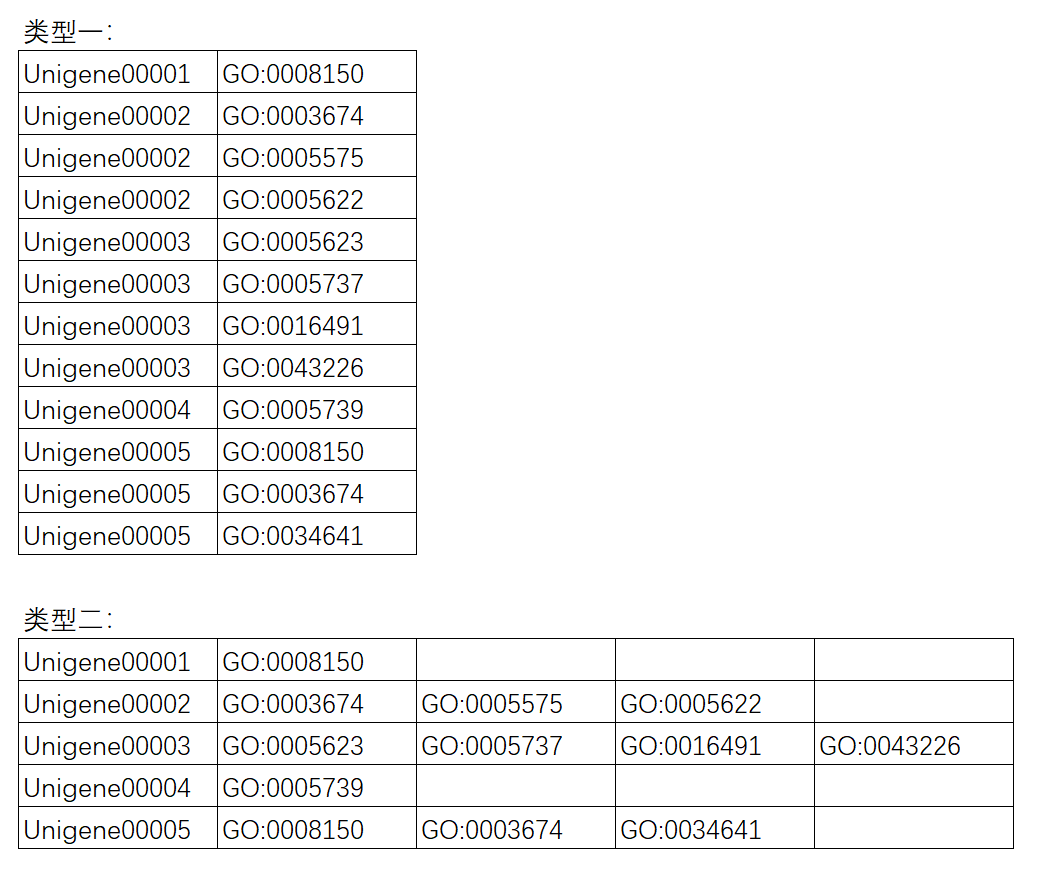
如果所研究物种不在以上范围,则需要自行准备GO背景基因文件。现在支持两种格式。第一种:格式为第一列为基因id,第二列为GO注释结果。第二种:同一个基因的所有GO号会在同一行并列给出。任务提交后,程序会自动判断处理。如下图所示:
输出:
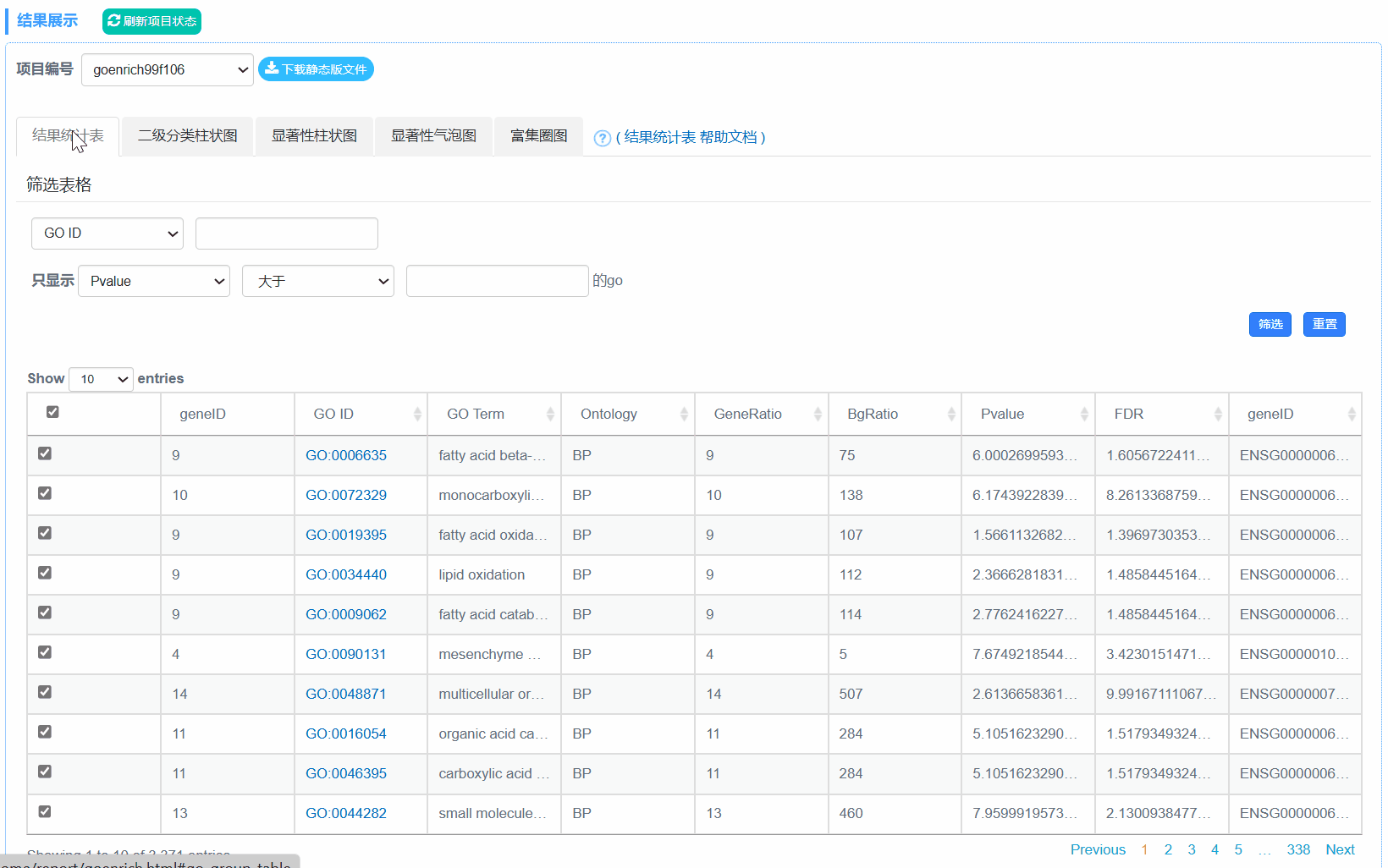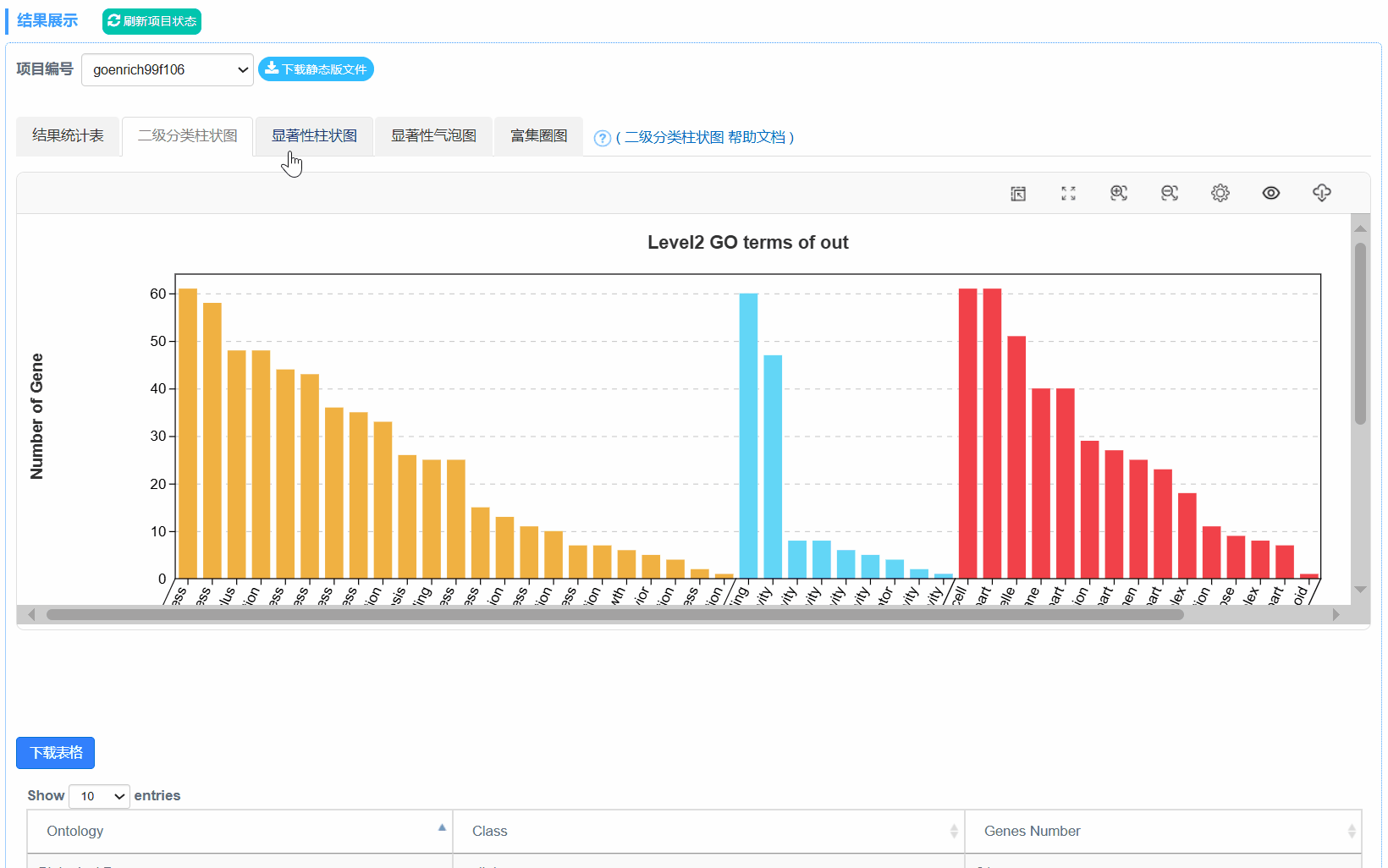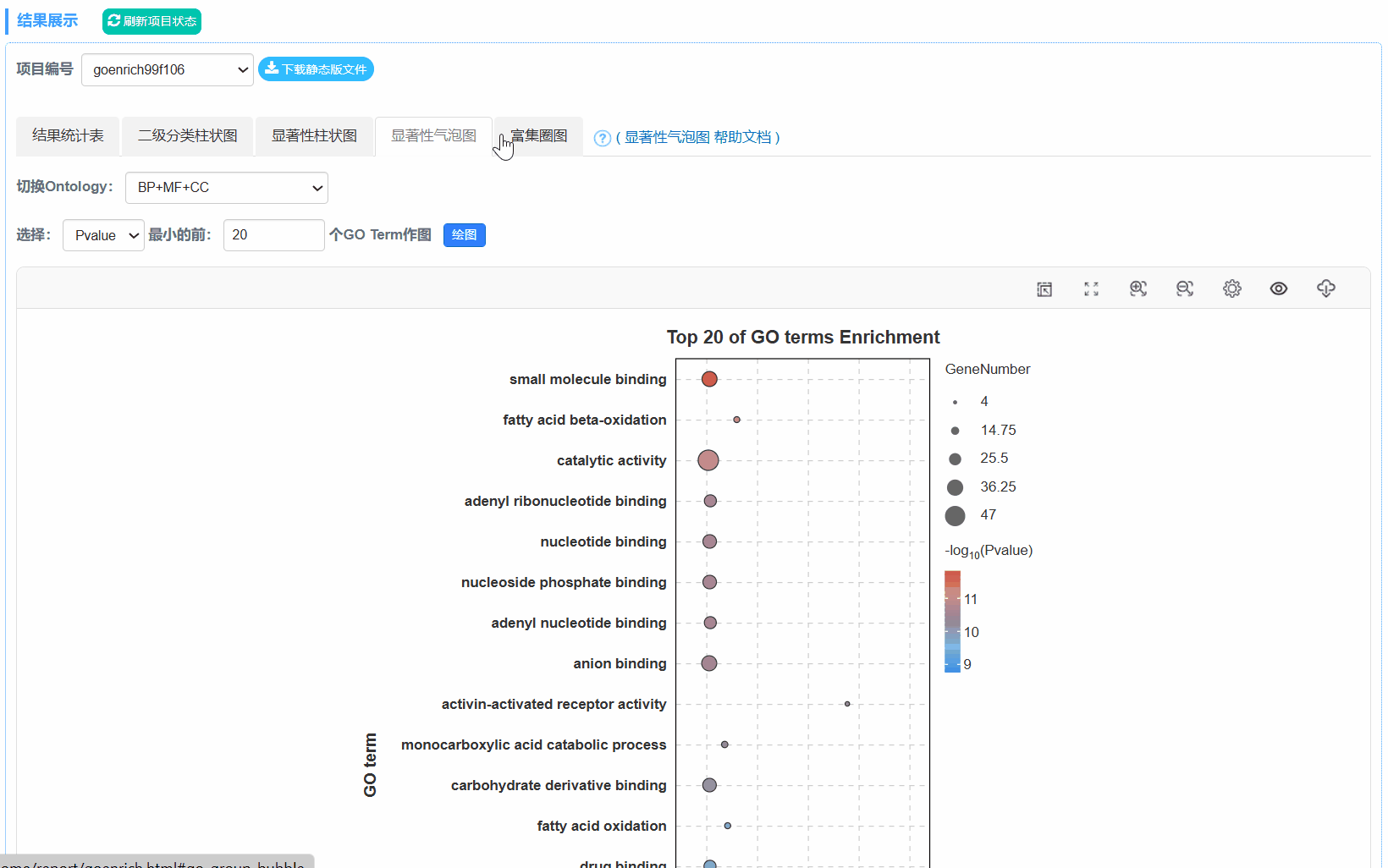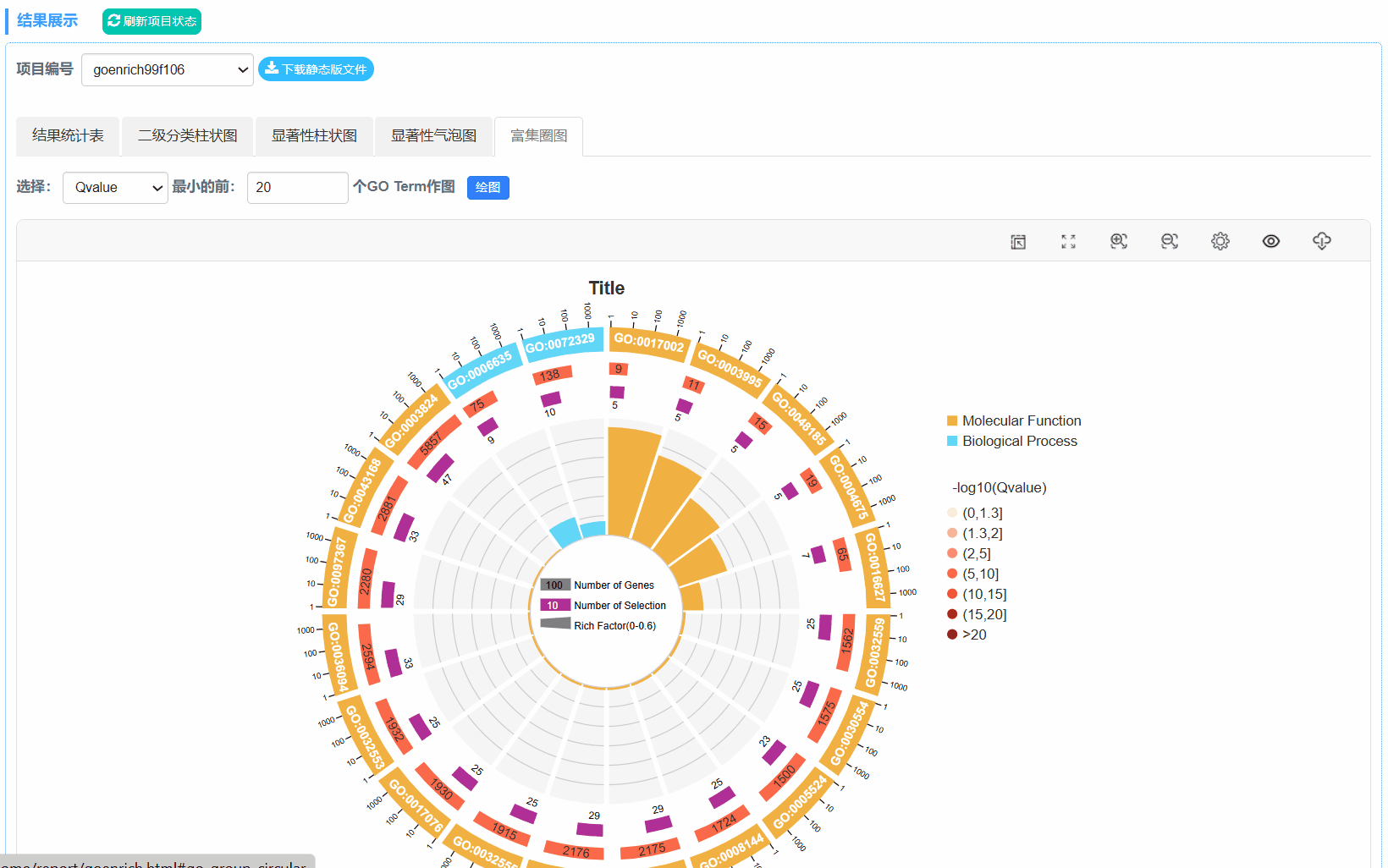
①out.[PFC].html: 网页格式结果,3个分别对应GO的3个主要分类。
②out.[PFC].xls: 基因的GO功能分类统计结果。
③out.[PFC].png:基因的GO功能分类结果统计图
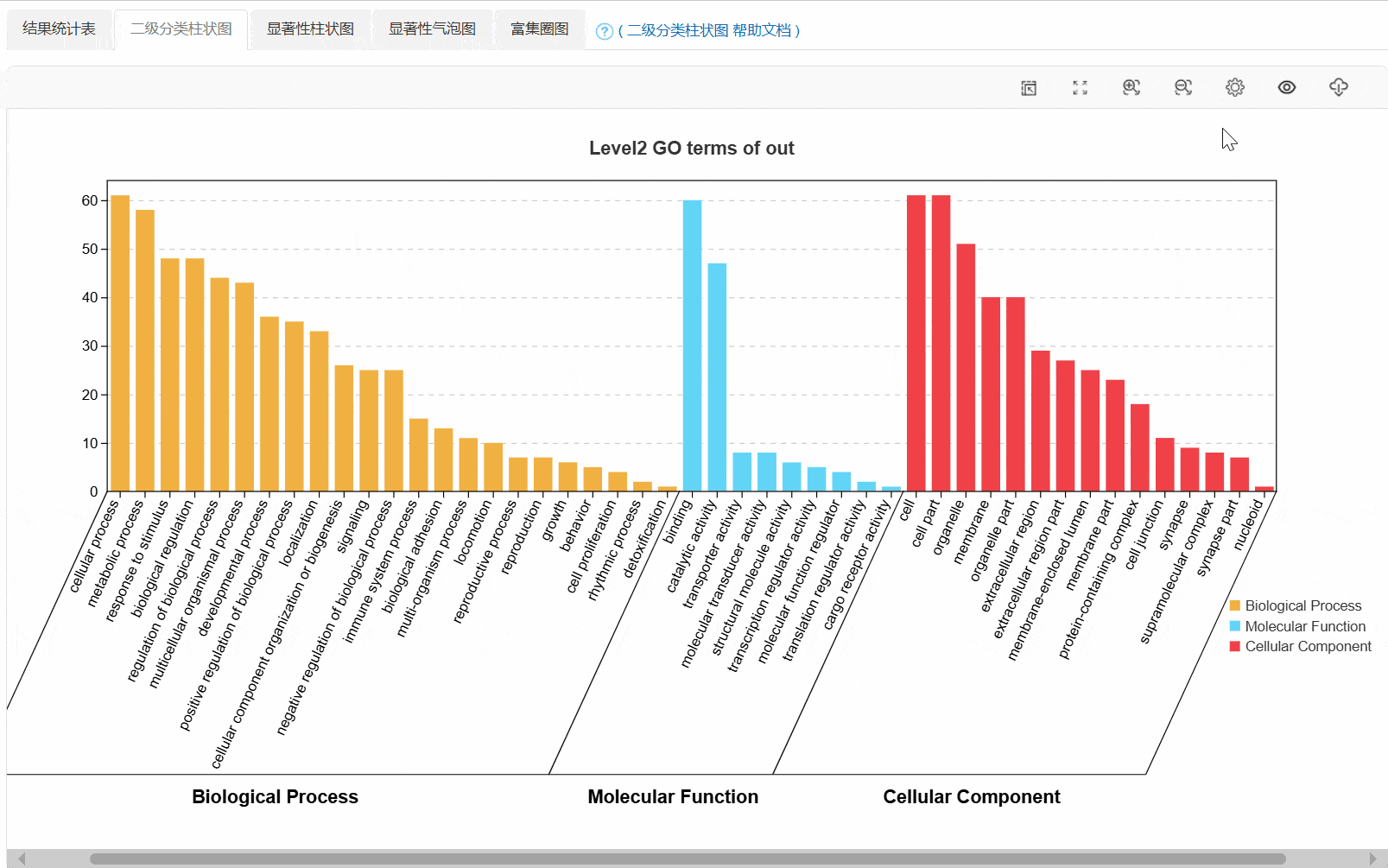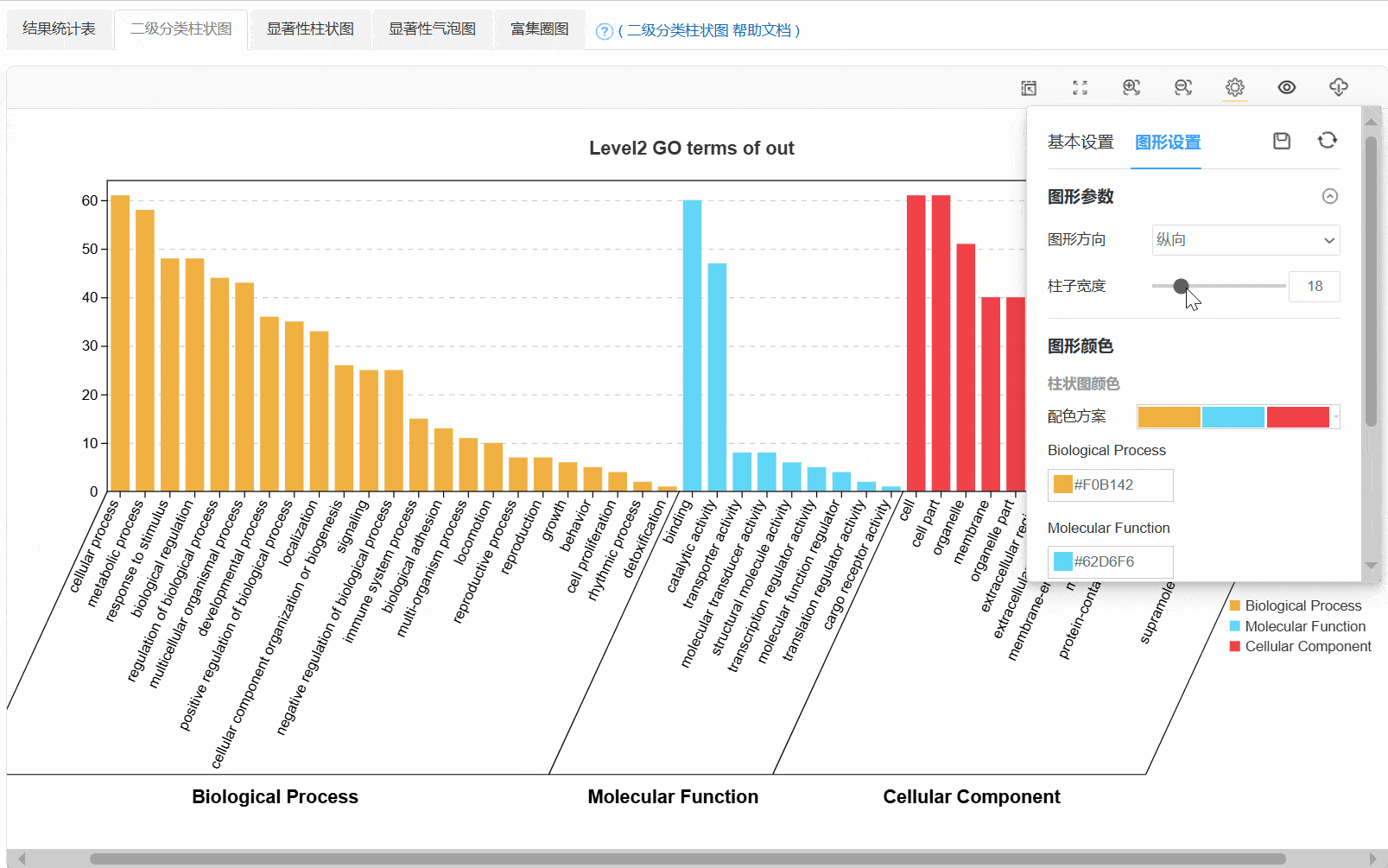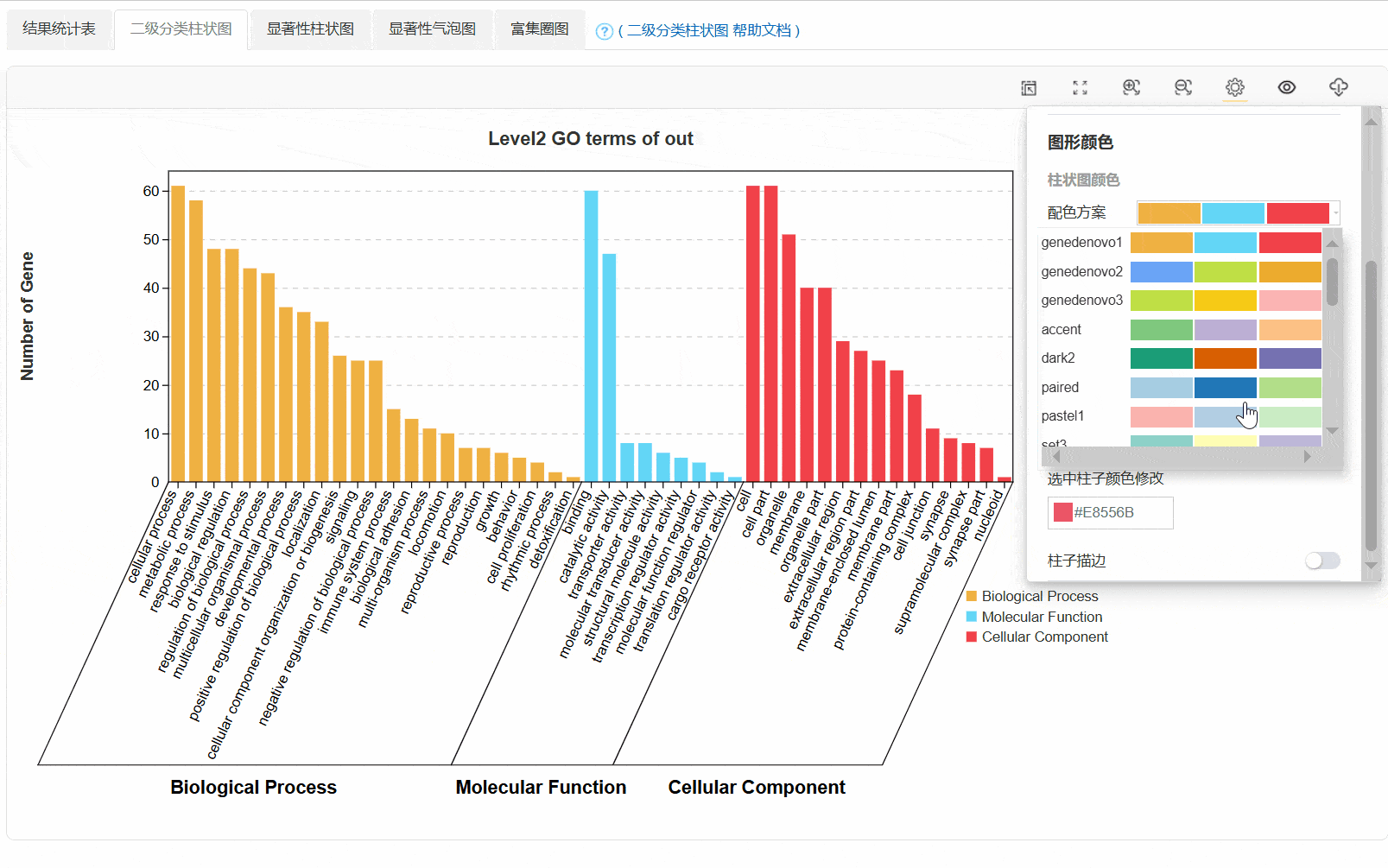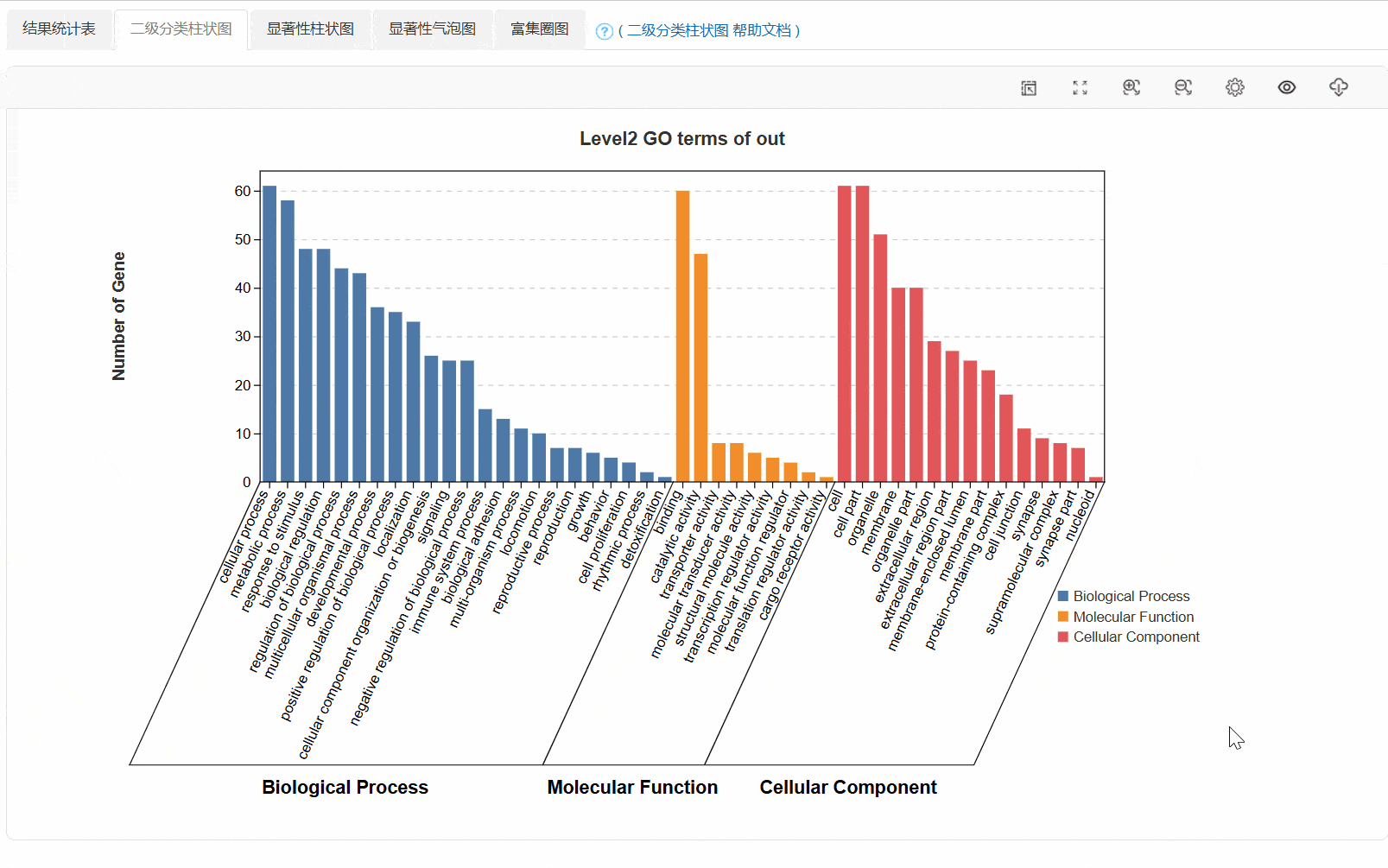
④out.secLevel2.svg/png:为GO二级分类统计图,统计了用于富集的基因在GO 的二级分类中占各个分类的数量,统计结果在xls表中。表格内容包括,Ontology,Class(GO的二级分类),基因数,具体基因id。
⑤out.level2.xls: GO第二级分类统计
1. 总览
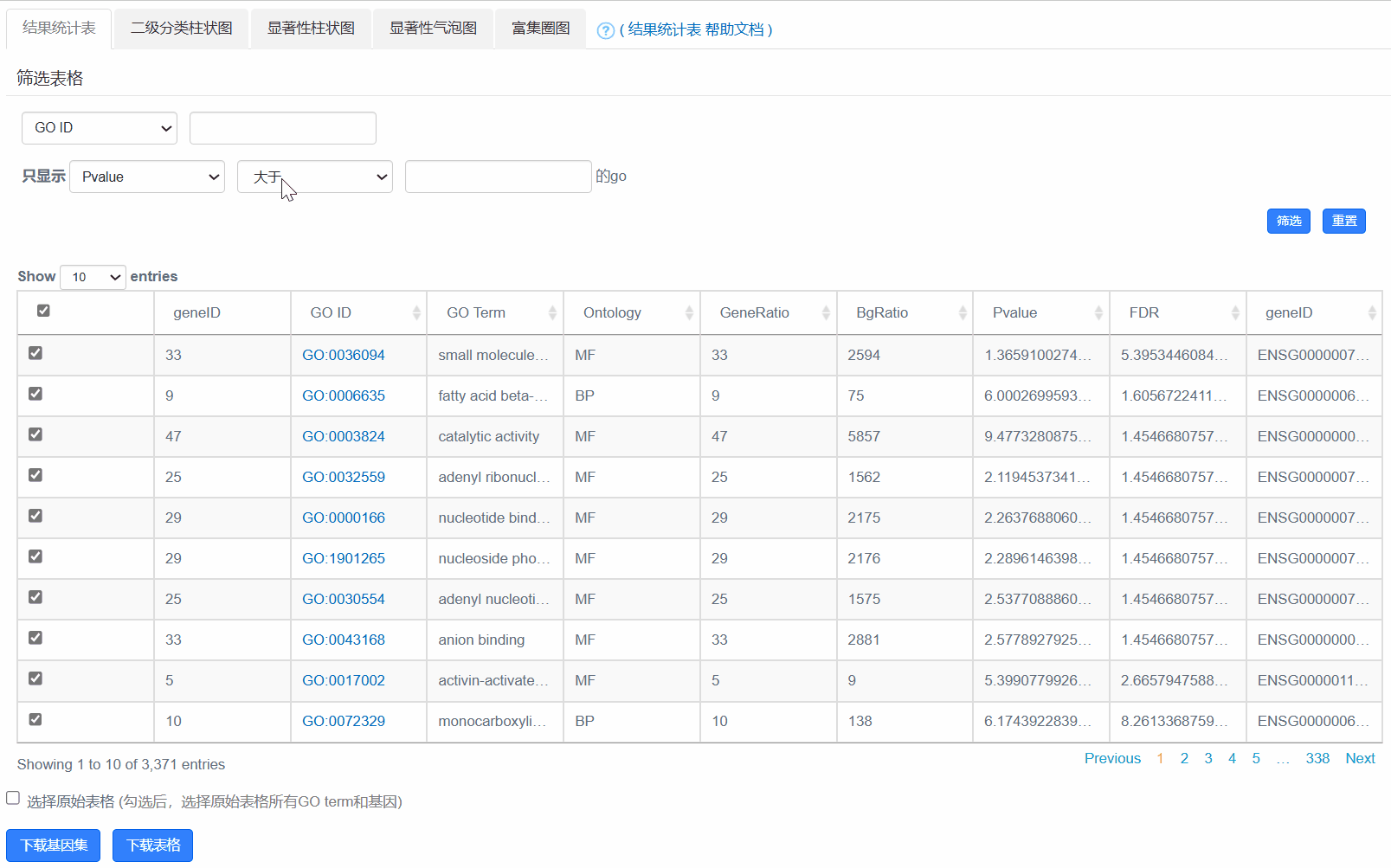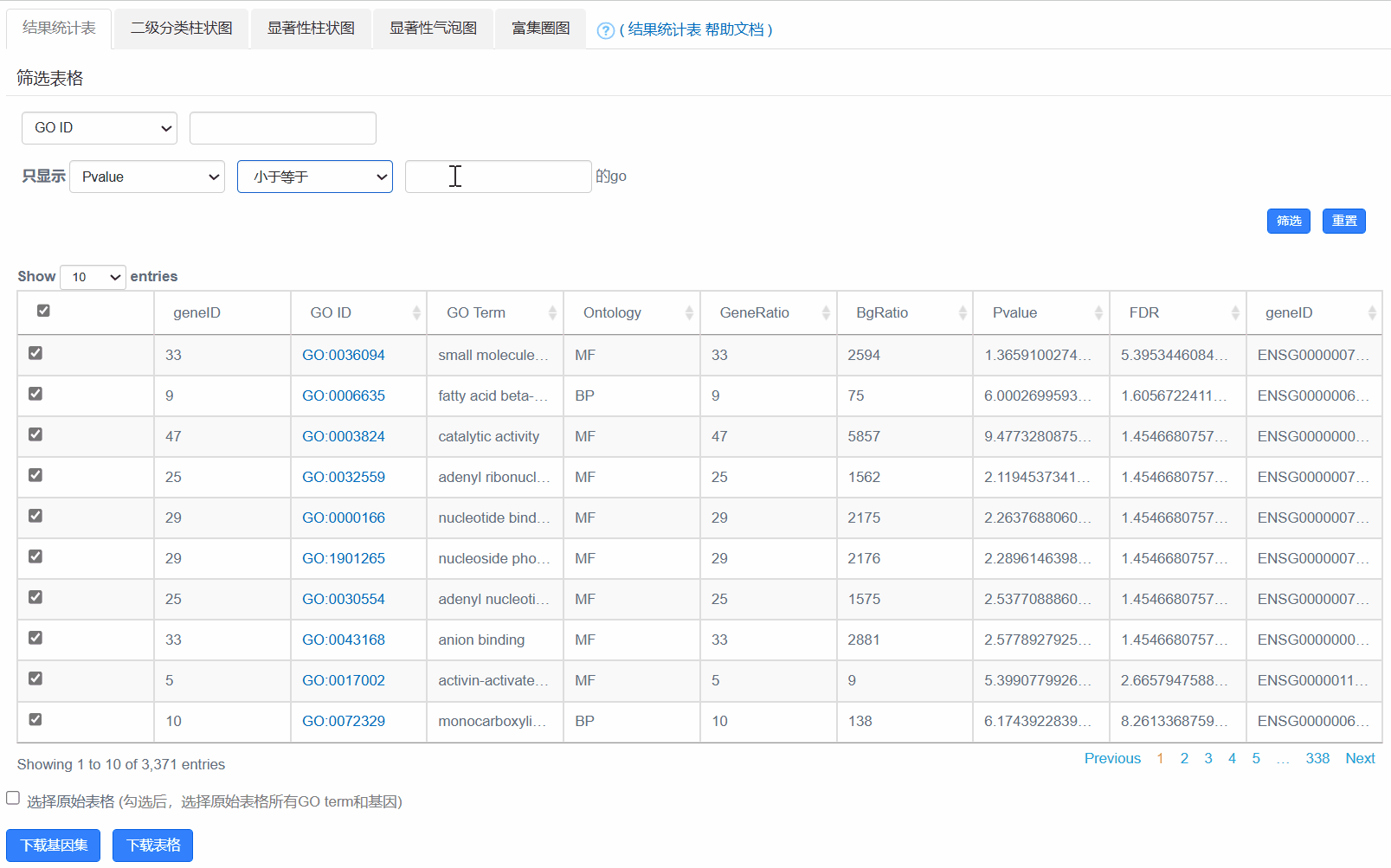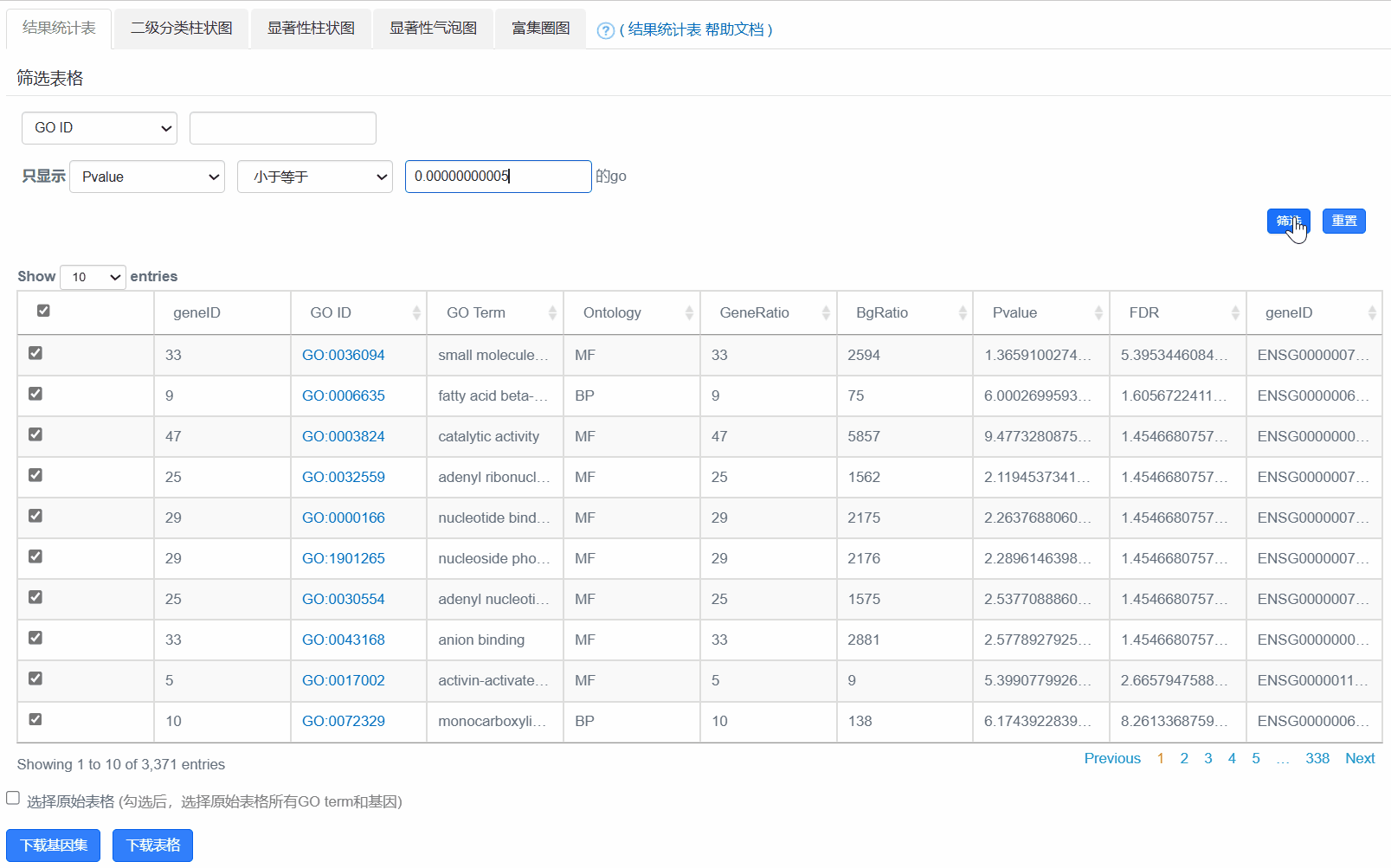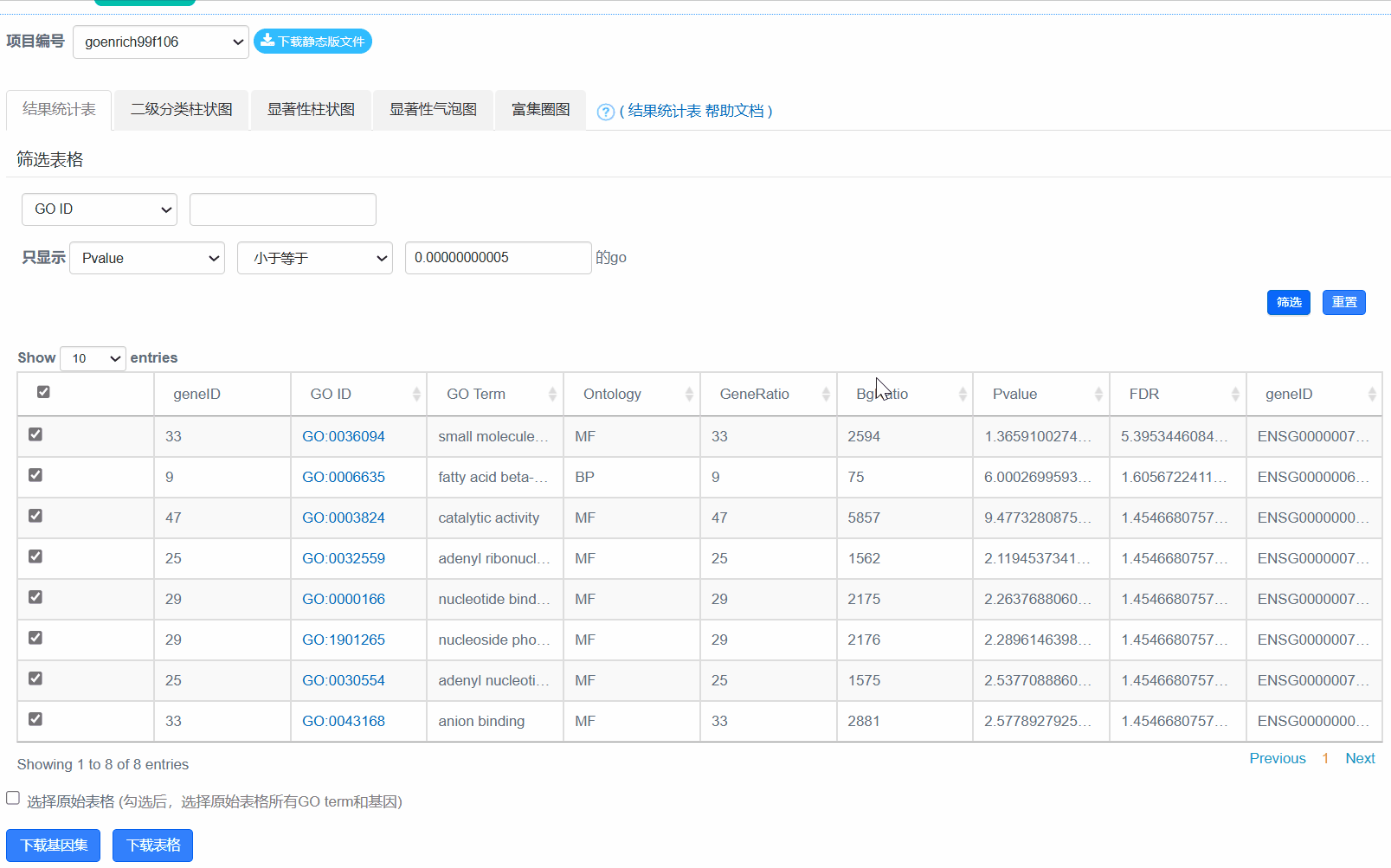
2. 结果统计表
3. 二级分类柱状图
4. 显著性柱状图
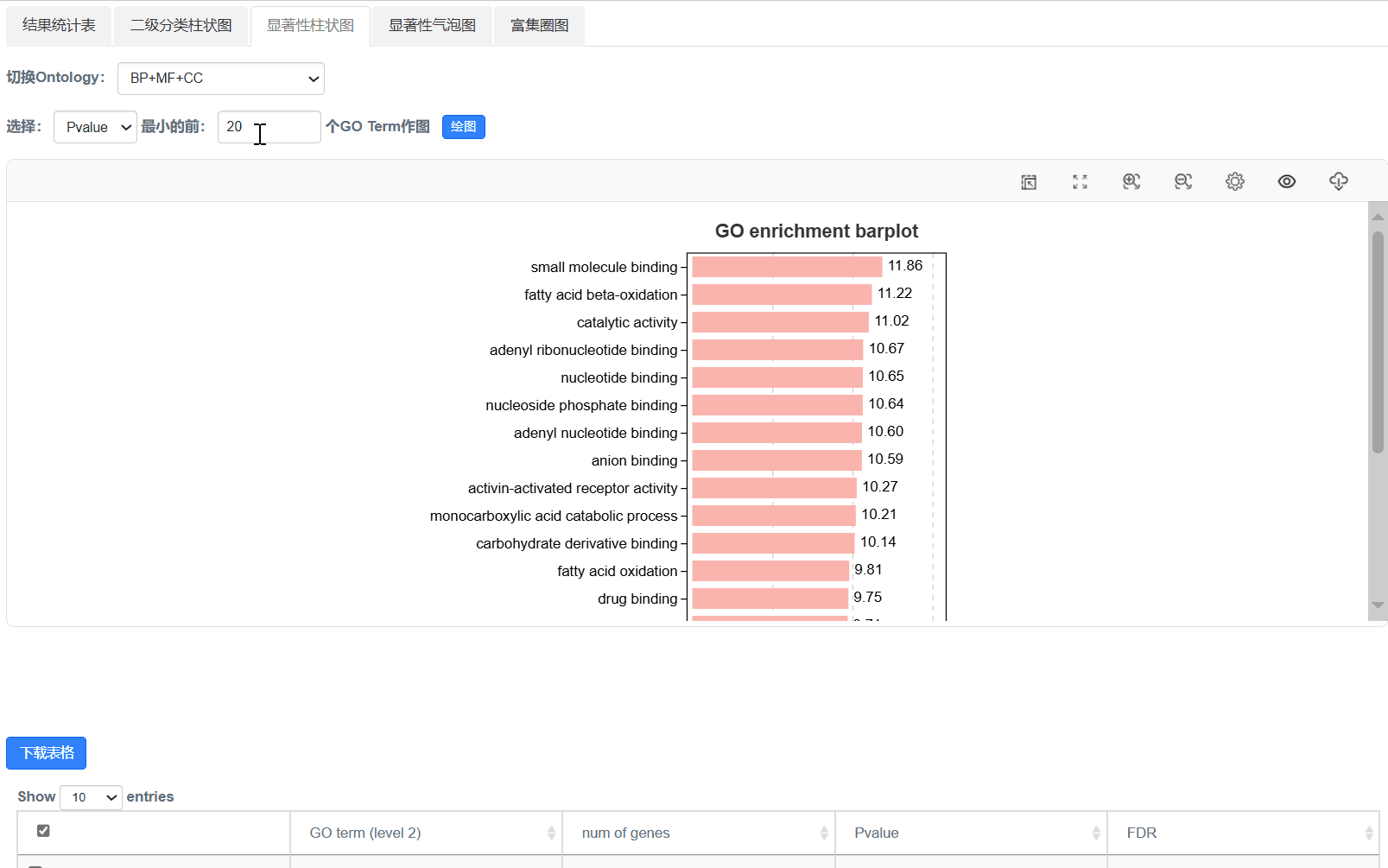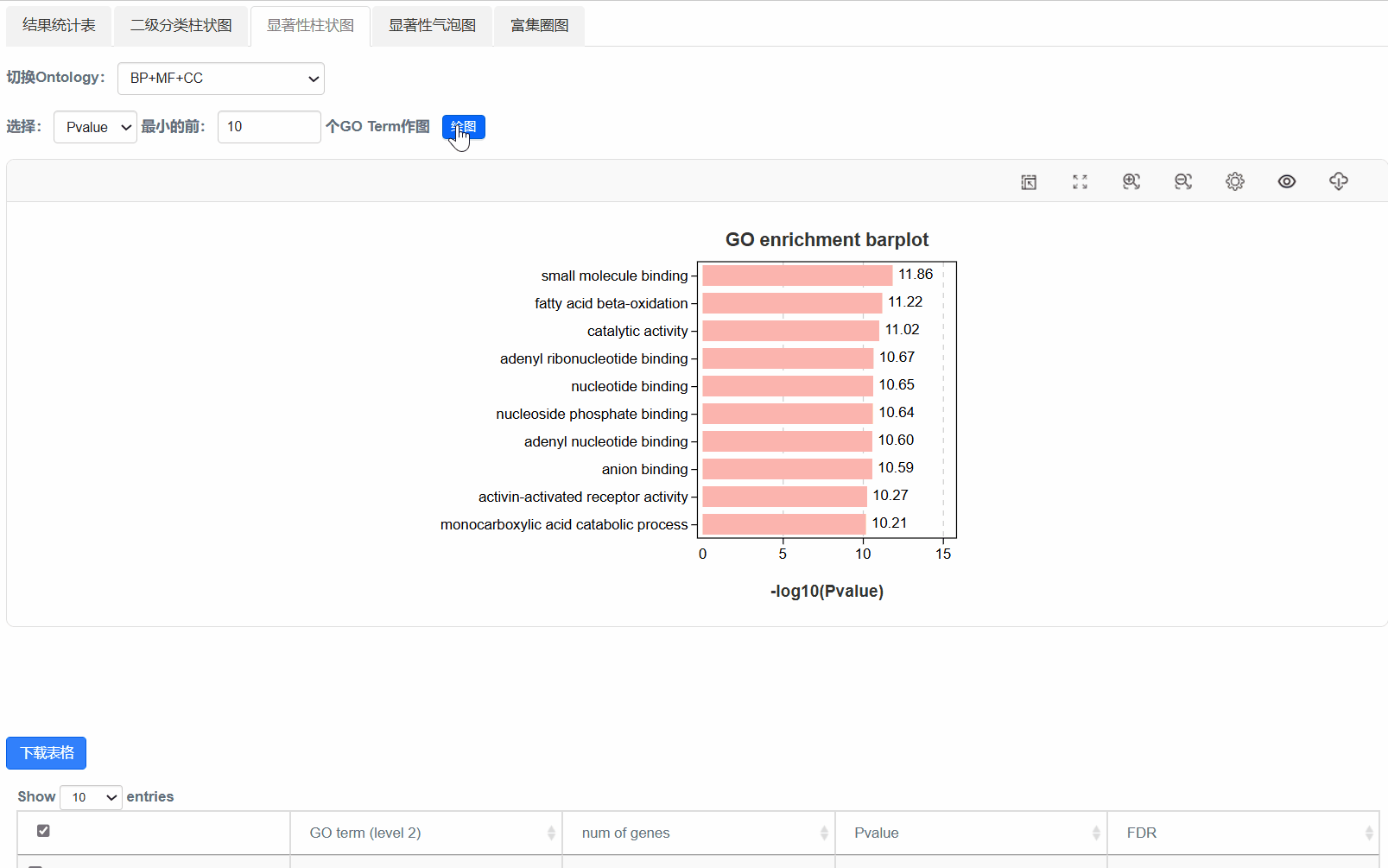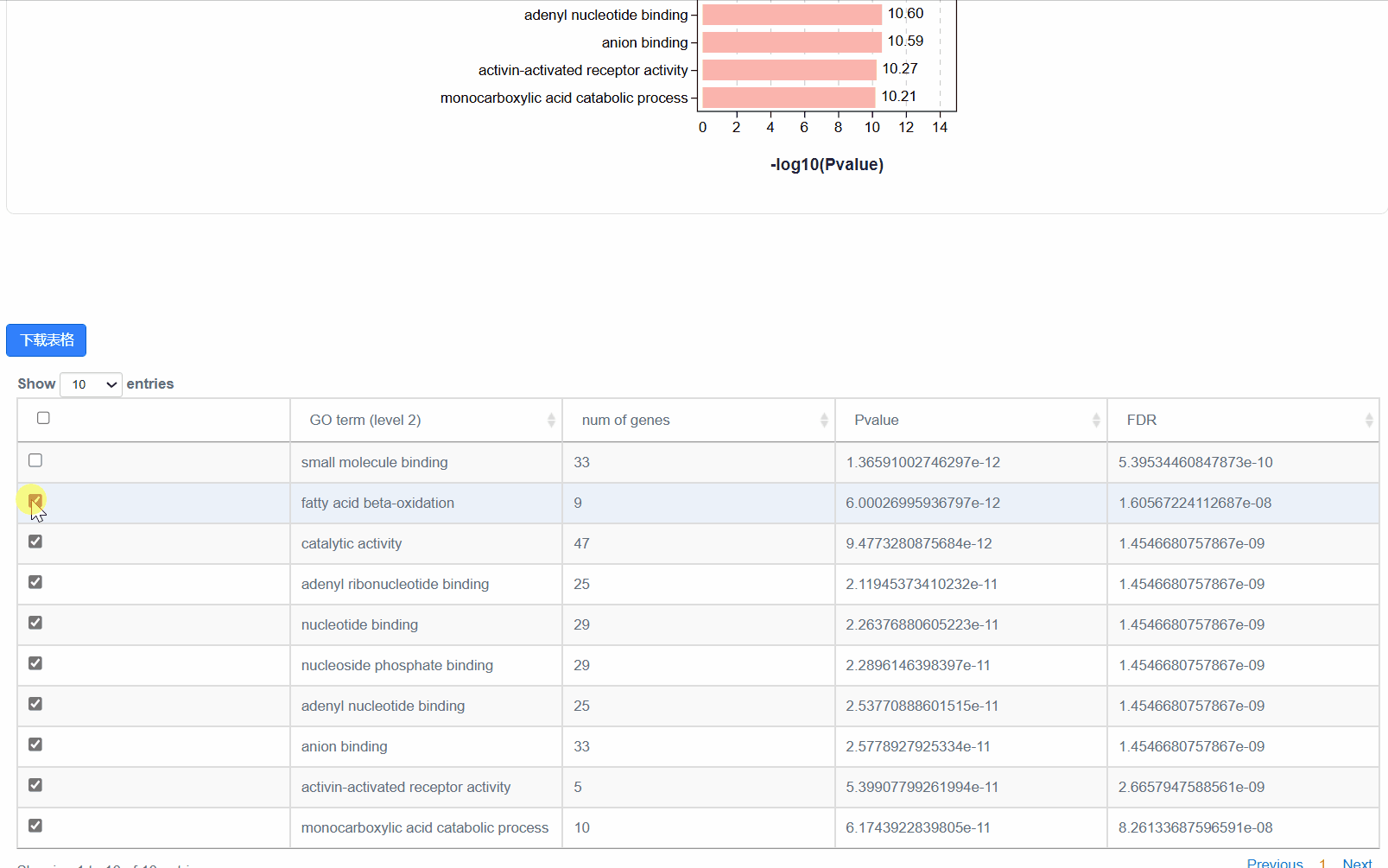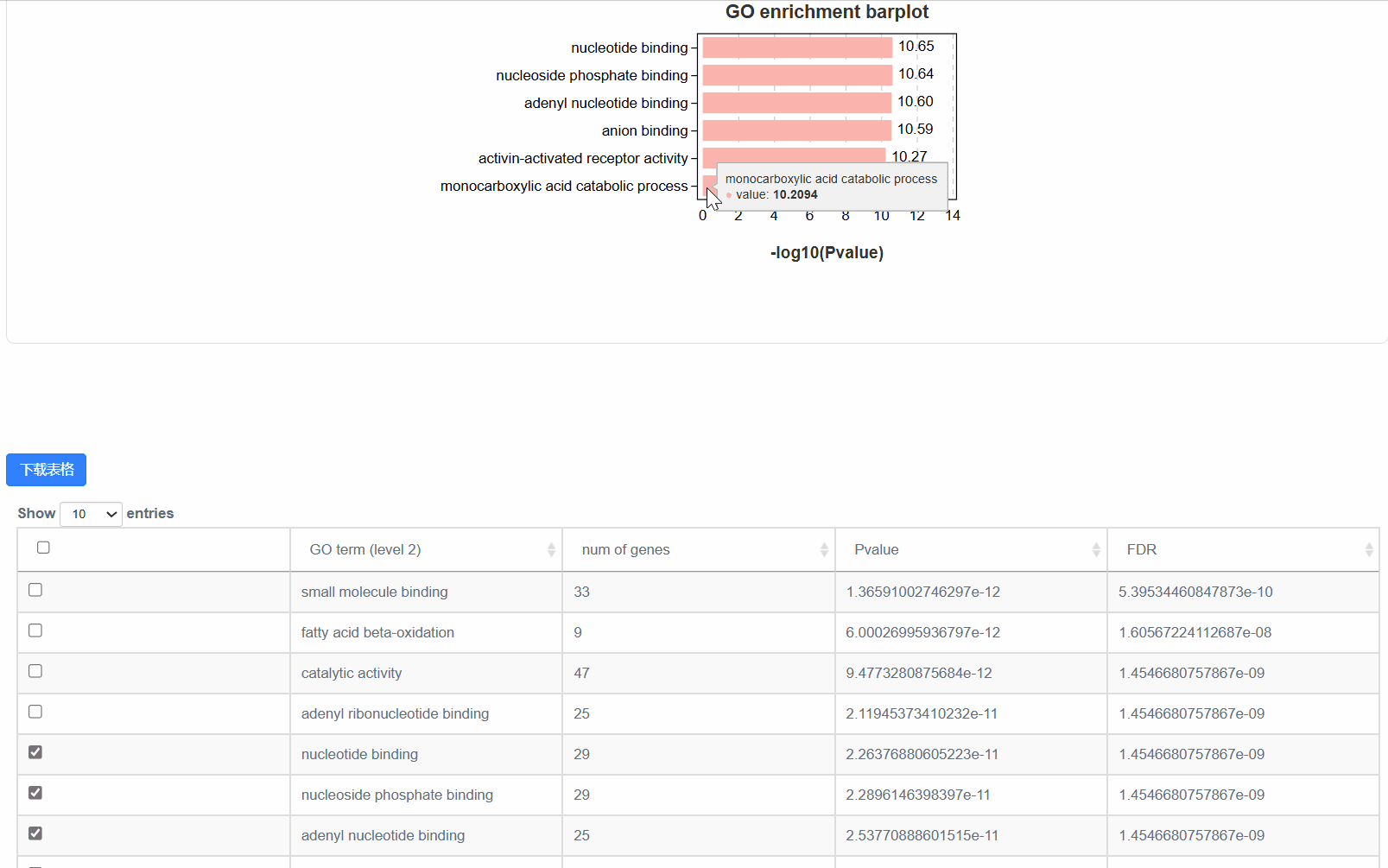
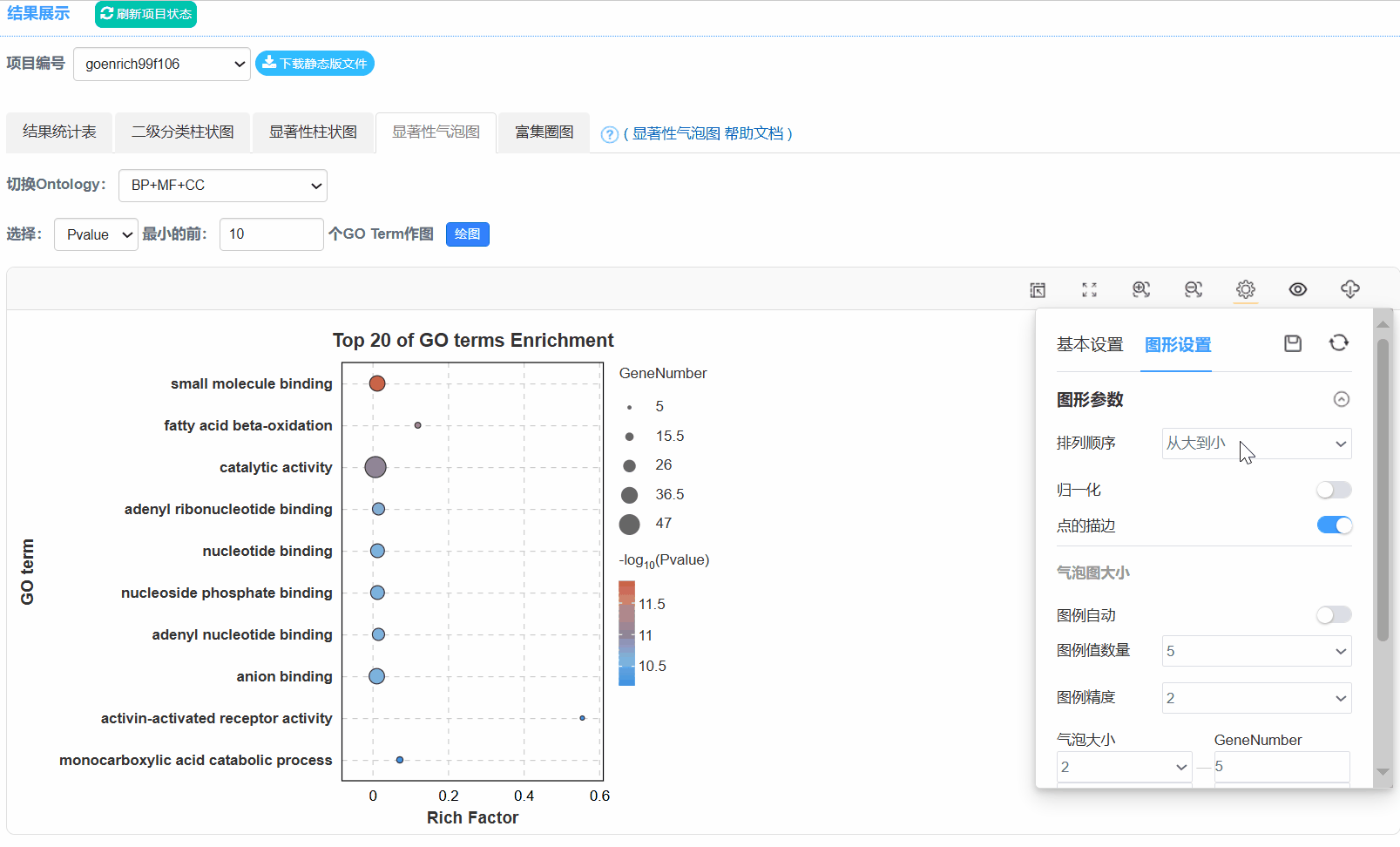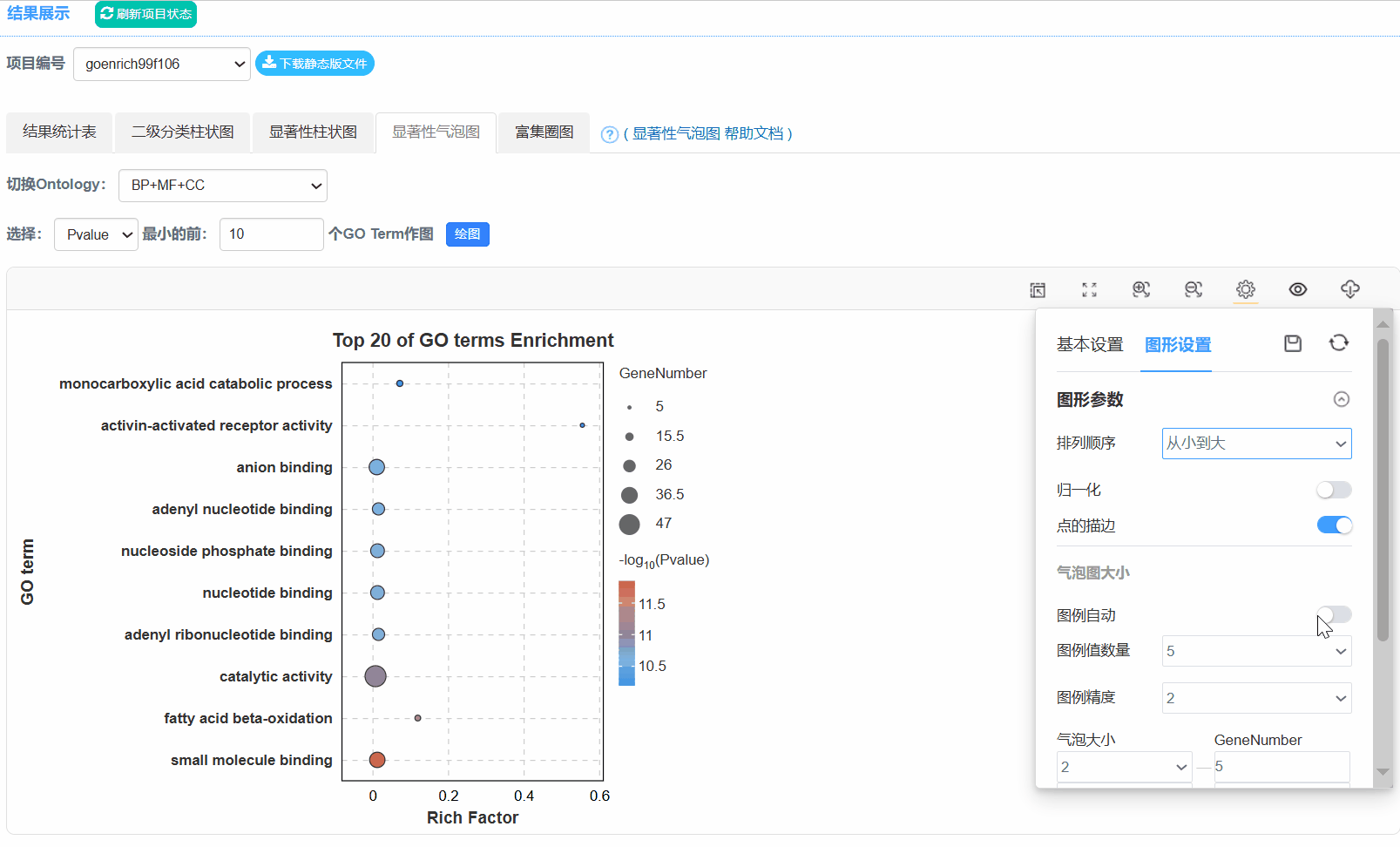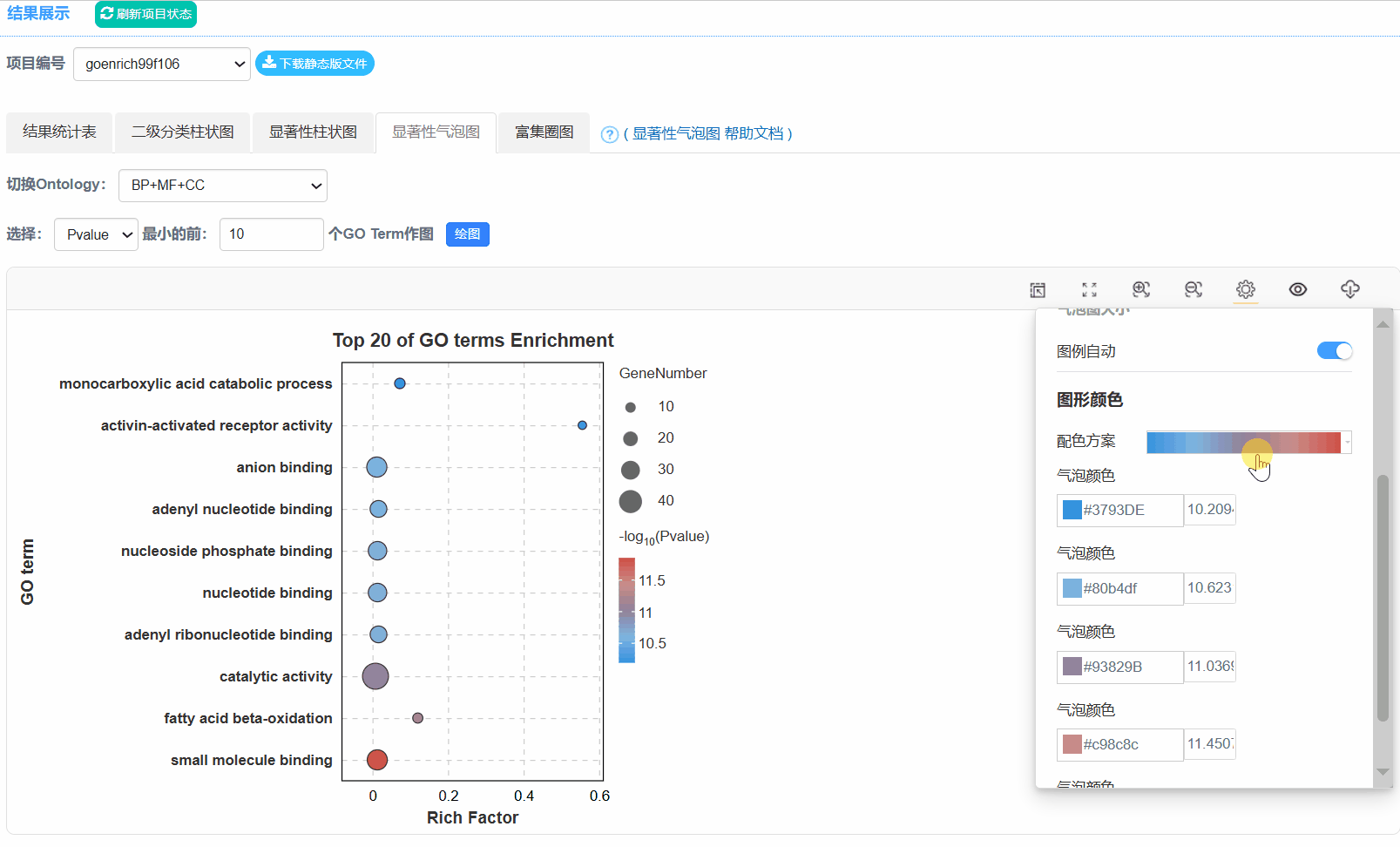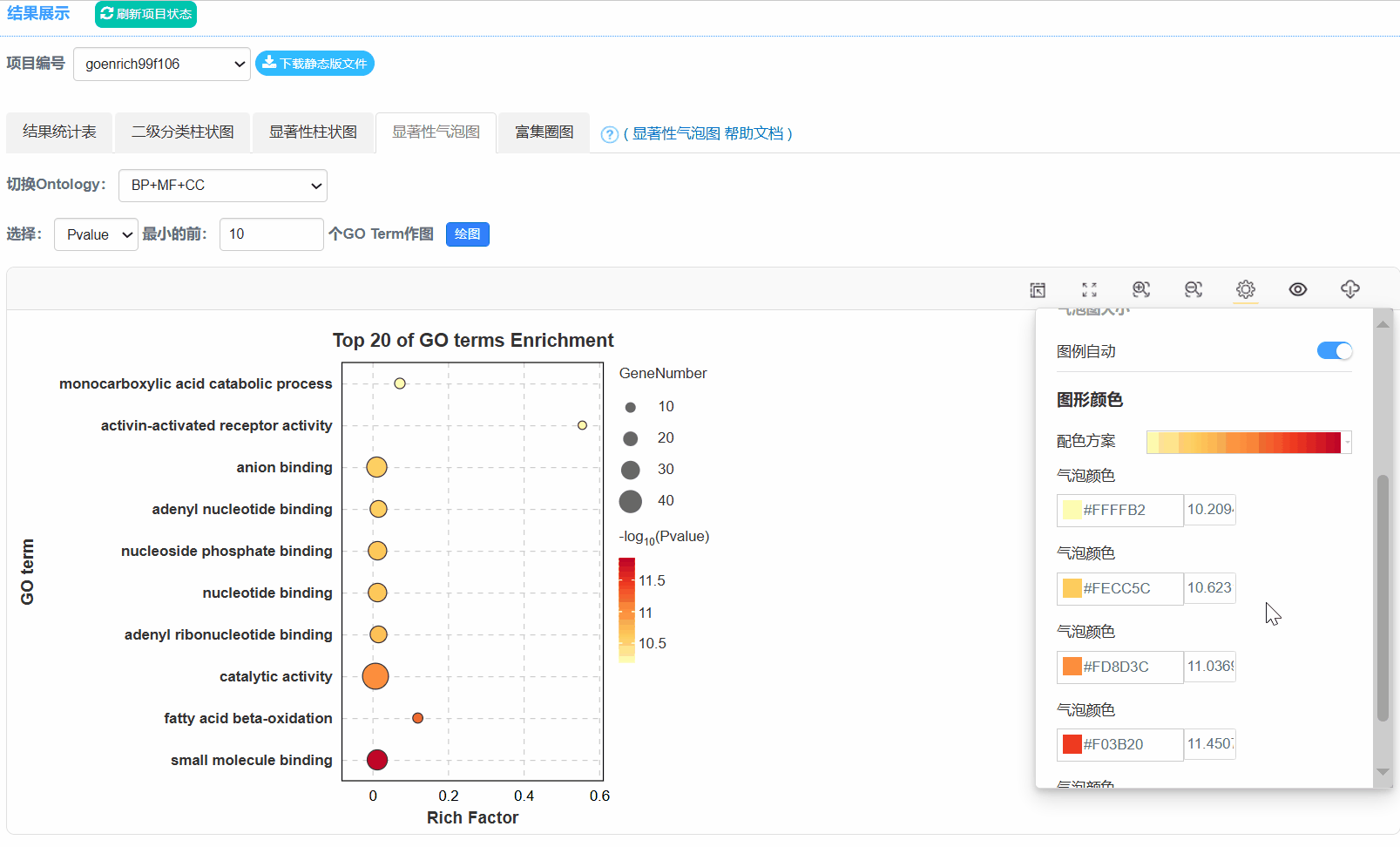
5. 显著性气泡图
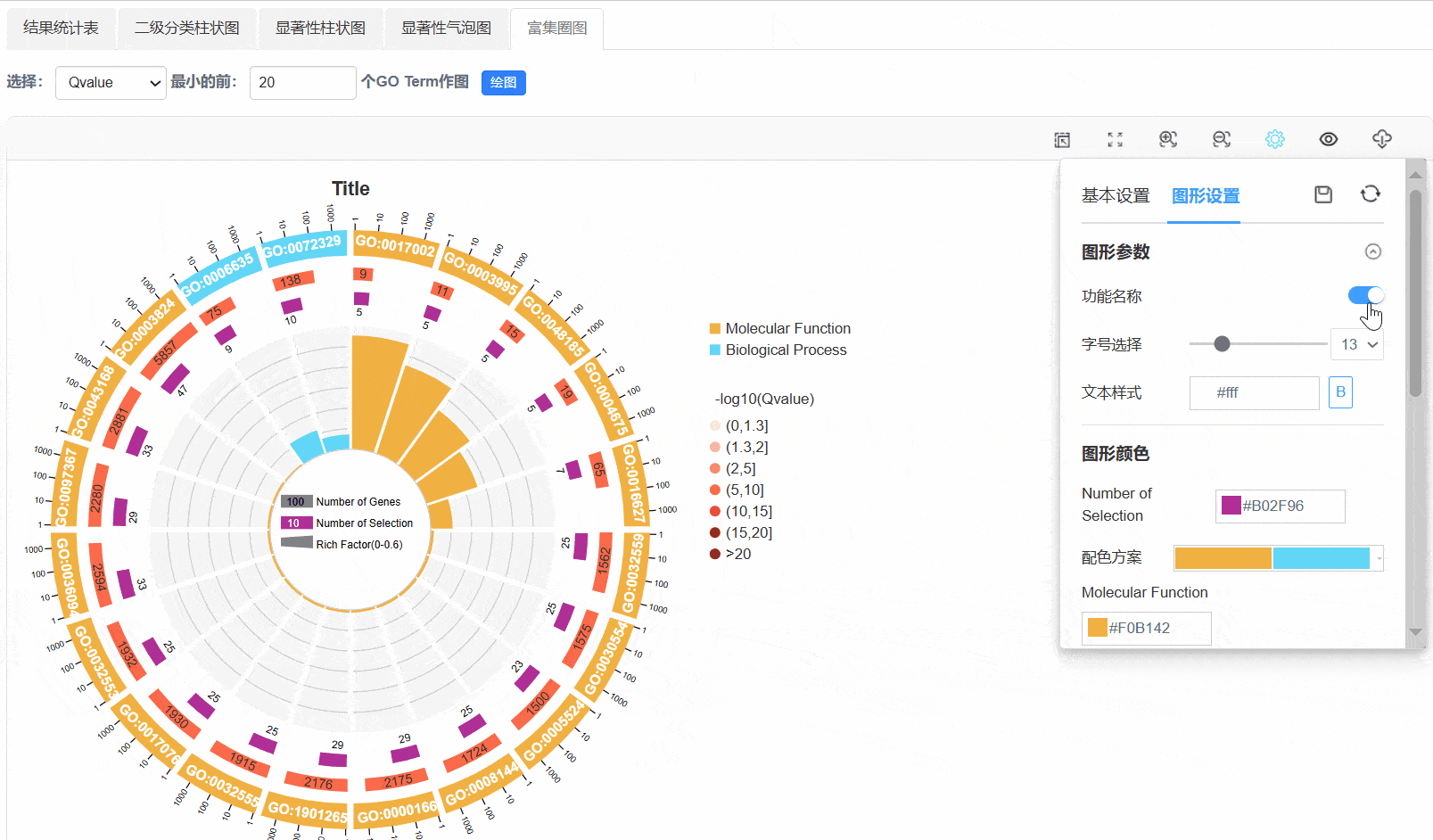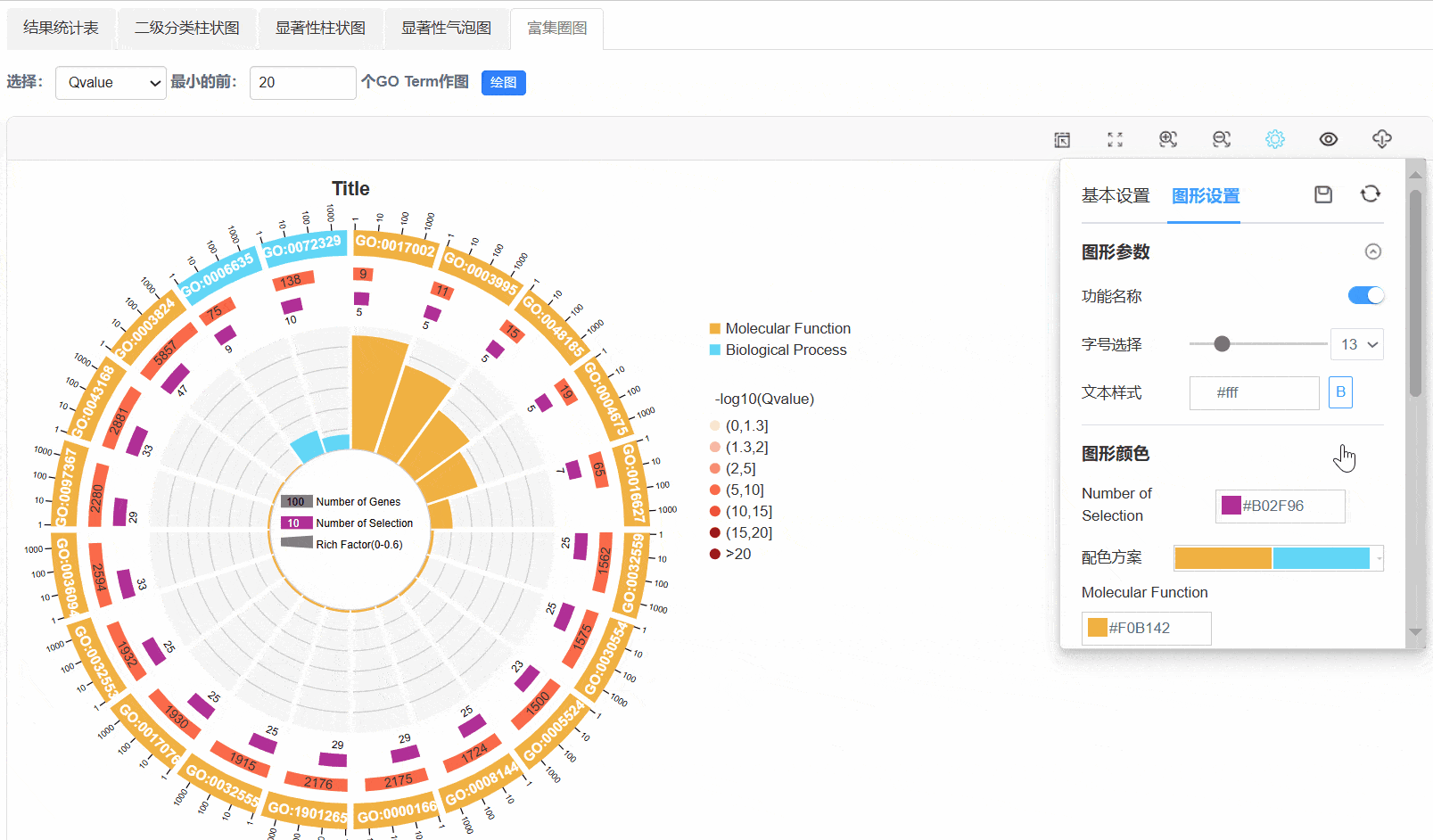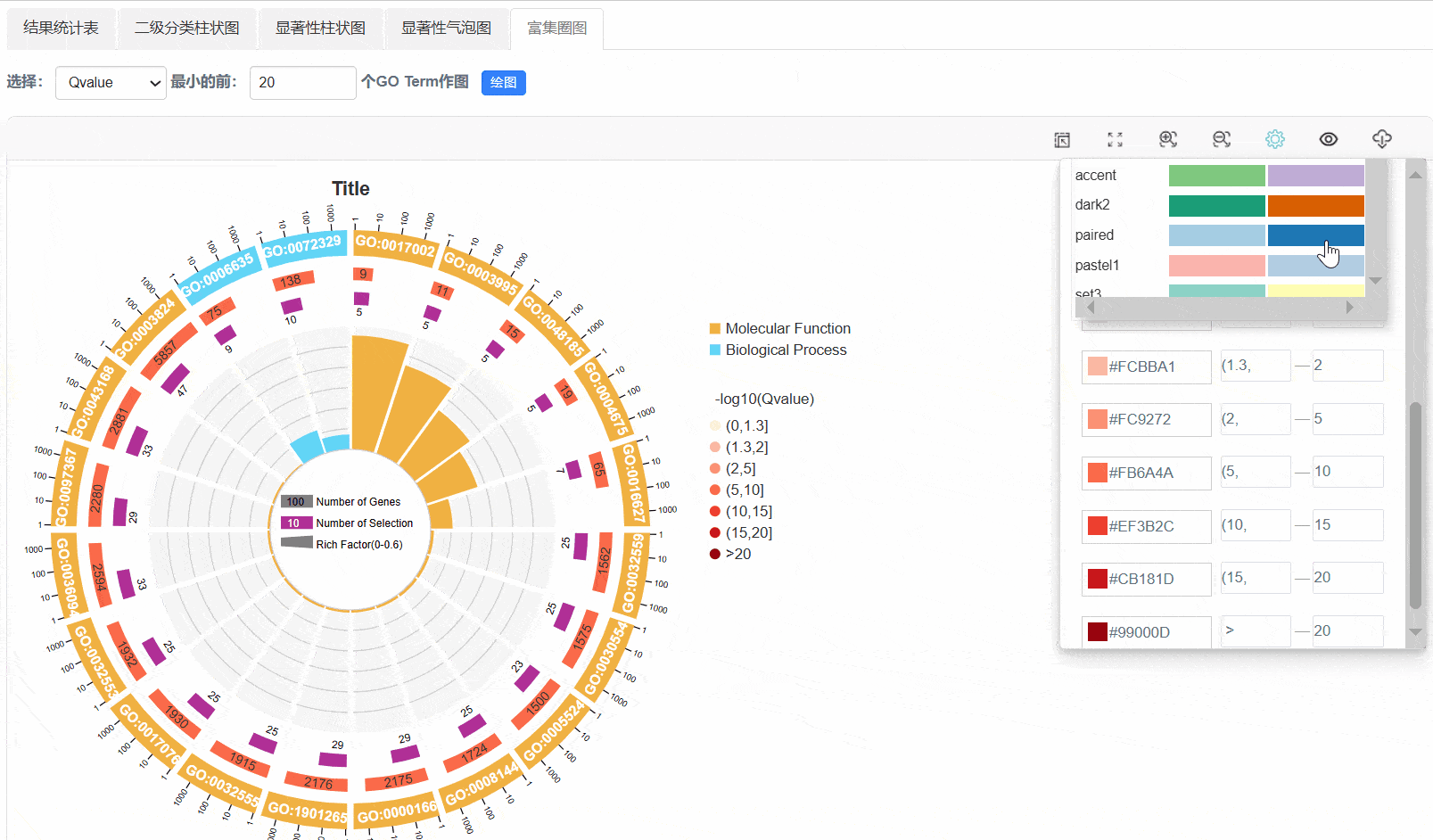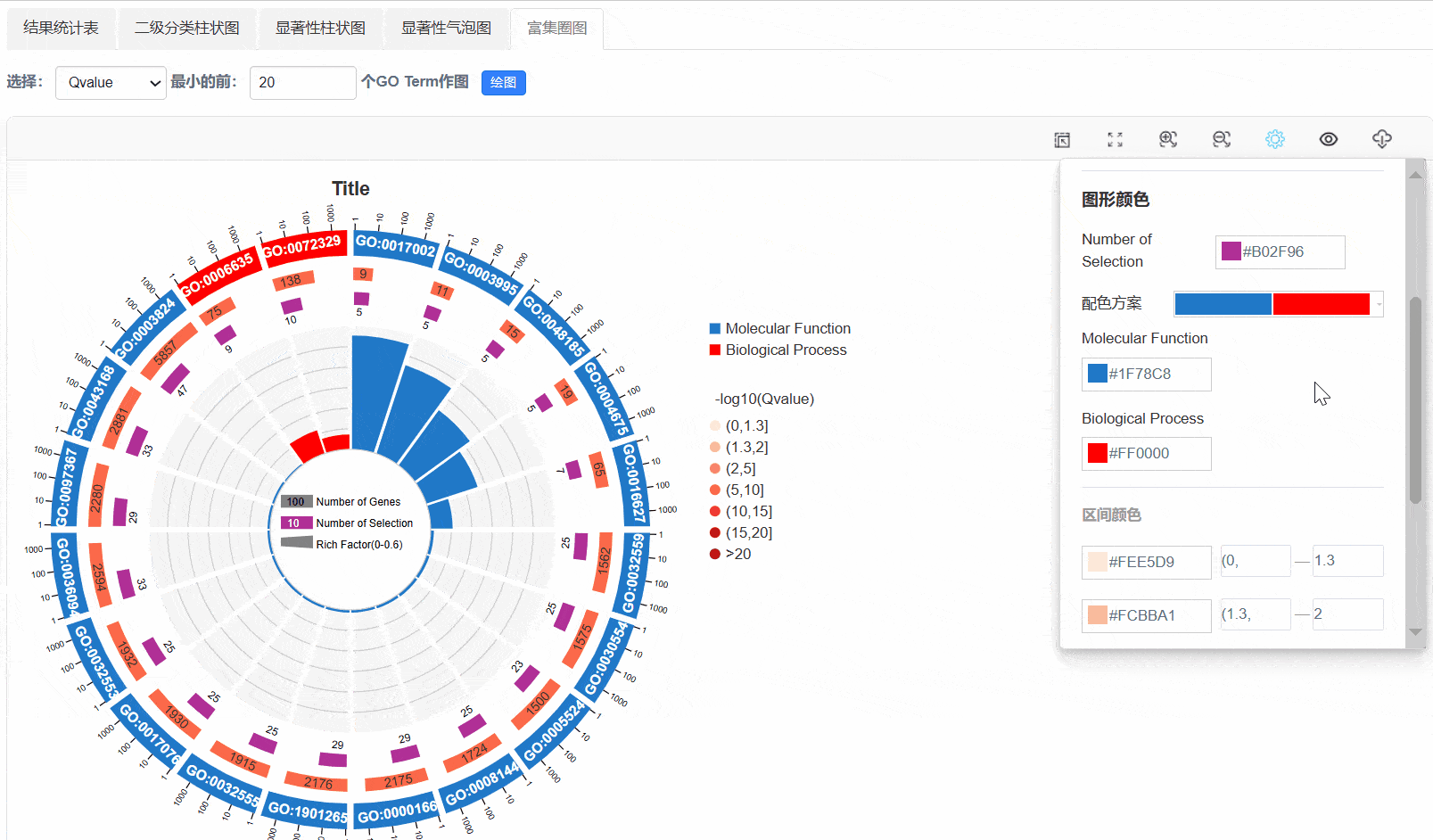
6. 富集圈图
Q1. 为什么上传目的基因,选用平台背景基因却出错?
(1)先确认自己物种是否已经切换。
(2)点击“预览背景基因”,查看平台提供的基因跟目的基因类型是否相同。
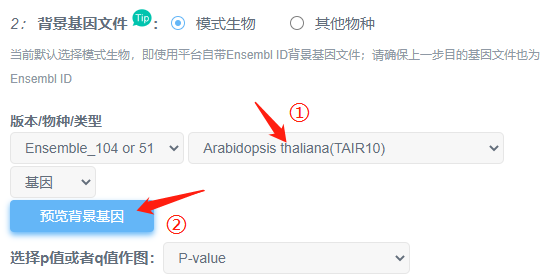
①平台提供的是Ensembl ID,但是目的基因表是symbol或其他id类型,则需要对目的基因进行基因ID转换;
②Ensembl id的结构是“物种前缀+序列类型+数字”Ensembl ID 后面的”小数”部分为版本号,如ENSG00000121410.11,小数部分的版本号必须删除。
Q2.怎么将基因转换成跟背景基因适配的类型呢?
①如果是上述18种常见物种,且手头上的基因类型是Gene_stable_ID/ Gene_name/NCBI_gene_ID/Gene_Synonym,那么可以使用“基因ID转换”(点击跳转)工具直接进行转换。
②如果不是上述物种或类型,可以通过BioMart等网址进行转换,相关教程在OmicShare论坛有很多,点击跳转相关教程。
【关于结果的常见问题】
Q3.结果文件中的P/F/C是什么意思?
分别对应BP/MF/CC。
GO总共有三个ontology(本体),分别描述基因的分子功能(molecular function)、细胞组分(cellular component)、参与的生物过程(biological process)
Q4.为什么二级分类统计图没有上下调基因统计?
文件中没有log2FC或者文件中有数据但上传时没有勾选“包含”log2FC列。
Q5.为什么结果中 p value 全为1:
pvalue全为1,基本是目的基因数目与背景基因数目完全一样所致。注意,富集分析中的背景基因是当前物种所有基因的列表。
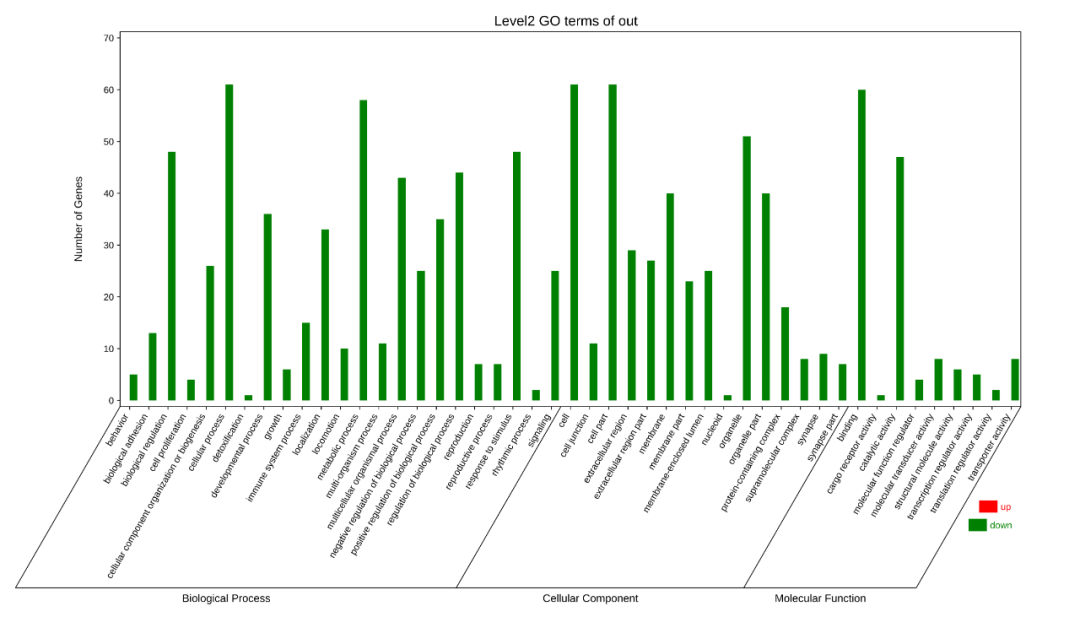
Q6.为什么二级分类统计图不是按照Gene number排序?
上传时,文件没有log2FC但勾选了“包含”,会导致出图时候基因数目没有按照降序排序,如下图。
Q7.为什么统计结果会比我上传的目的基因数目多?
由于一个基因常常对应多个GO term,因此同一个基因会在不同分类条目下出现,即被多次统计,因此如果把二级分类统计图所有柱子的基因数目加起来,肯定是多于profiel1总的基因数目的。
引用OmicShare Tools的参考文献为:
Mu, Hongyan, Jianzhou Chen, Wenjie Huang, Gui Huang, Meiying Deng, Shimiao Hong, Peng Ai, Chuan Gao, and Huangkai Zhou. 2024. “OmicShare tools: a Zero‐Code Interactive Online Platform for Biological Data Analysis and Visualization.” iMeta e228. https://doi.org/10.1002/imt2.228案例1:
发表期刊:Signal Transduction and Targeted Therapy
影响因子:39.3
发表时间:2022
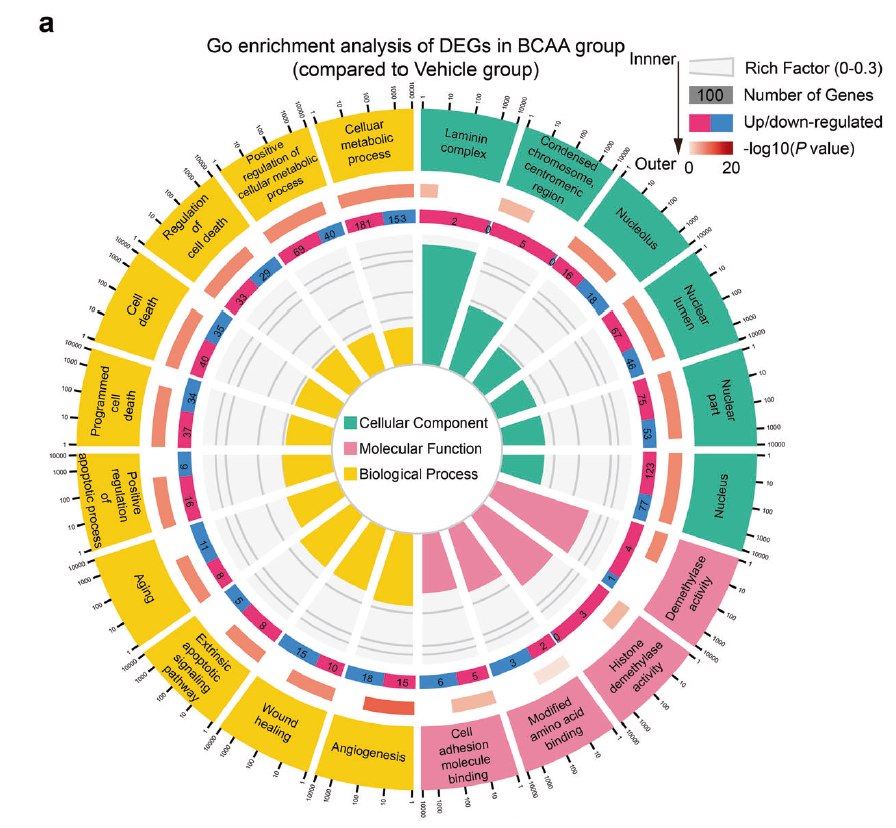
Fig. 3 BCAA accelerated H3K9me3 loss in ADSCs upon exposure to detrimental stress. a GO enrichment of the DEGs in ADSCs treated with vehicle or BCAA (3.432 mM) under hydrogen peroxide (100 μM) stress as identified by RNA-seq. Twenty significantly enriched GO terms are shown.
引用方式:
Gene Ontology (GO) enrichment analysis was performed and the results visualized with the
OmicShare tool, an online platform for data analysis (https://www.omicshare.com/tools/Home/Soft/enrich_circle).
参考文献:
Zhang F, Hu G, Chen X, et al. Excessive branched-chain amino acid accumulation restricts mesenchymal stem cell-based therapy efficacy in myocardial infarction[J]. Signal Transduction and Targeted Therapy, 2022, 7(1): 171-171.
案例2:
发表期刊:Nutrients
影响因子:5.9
发表时间:2022
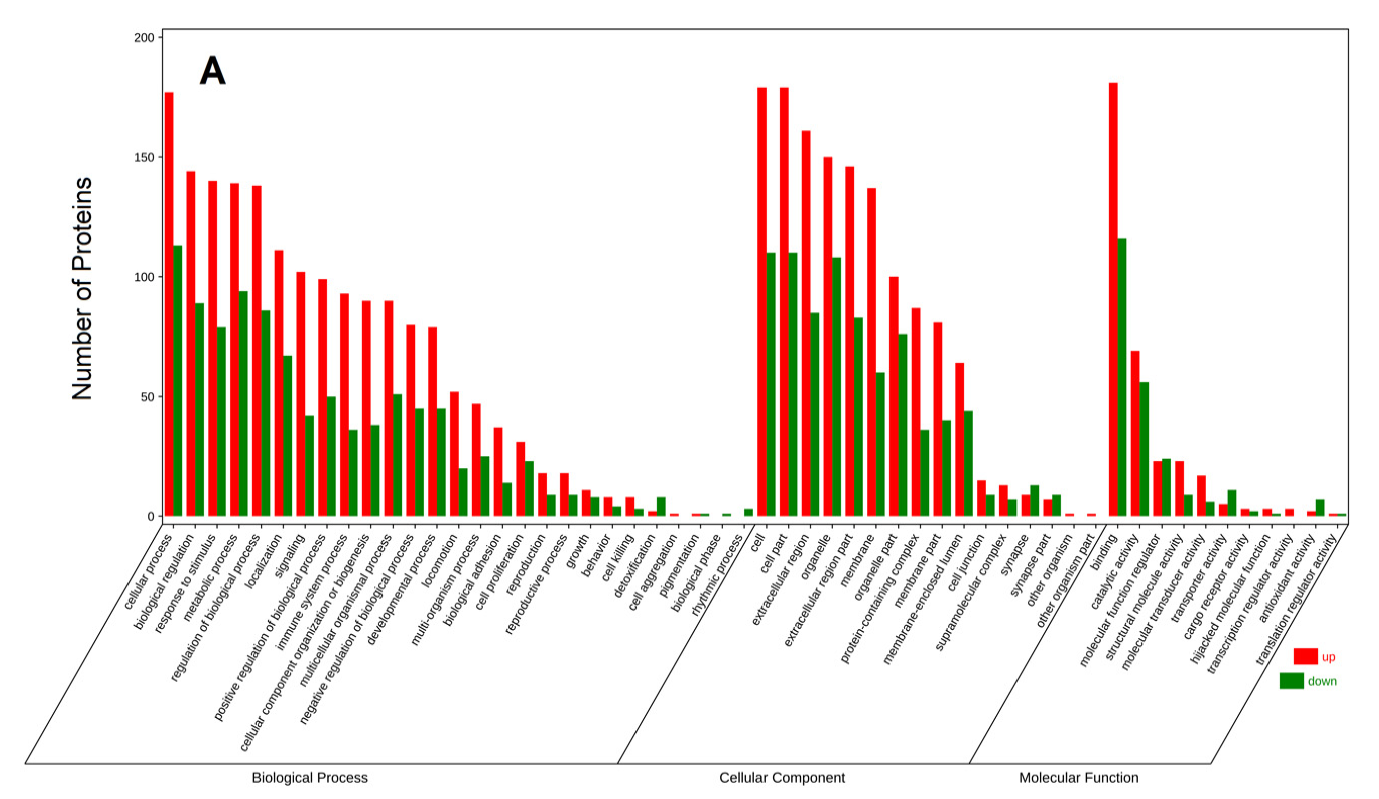
Figure 4. GO annotation of differentially expressed proteins in C vs. M(A)
引用方式:Omicshare online software was used for gene ontology (GO) annotation to analyze the annotation function of milk protein. Pathway analysis of the identified milk proteins was performed based on the online Omicshare software using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database.
参考文献:
Zhang Y, Zhang X, Mi L, et al. Comparative proteomic analysis of proteins in breast milk during different lactation periods[J]. Nutrients, 2022, 14(17): 3648.
案例3:
发表期刊:International Journal of Biological Macromolecules
影响因子:8.2
发表时间:2023
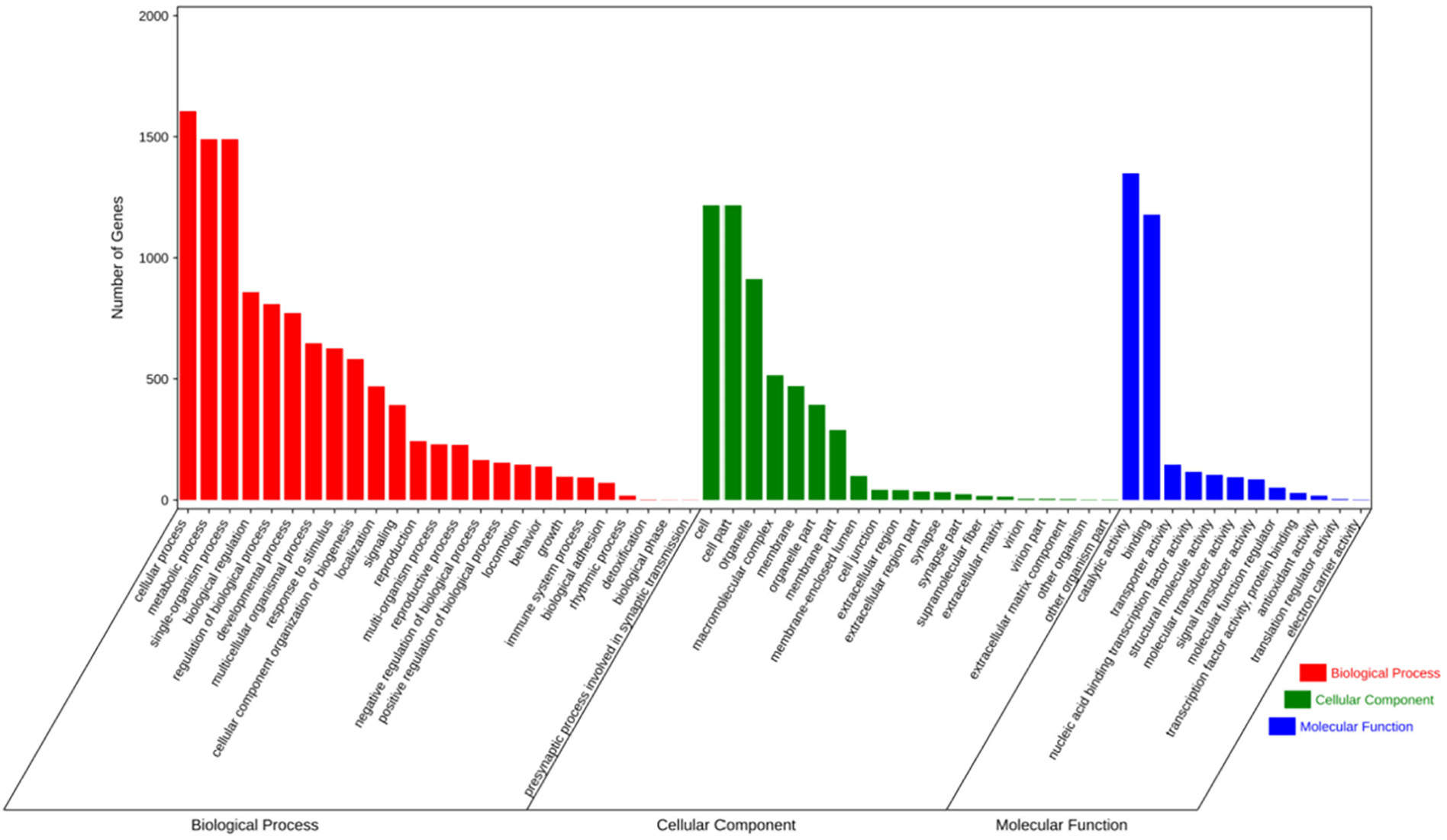
Fig. 2. GO enrichment analysis of G4 containing genes. The genes whose contained G4 were selected to do enrichment tests in order to reveal the enrichment of gene ontology terms. Statistically significant molecular functions, biological processes, and cellular component were identified. Each color block represents the gene amount in each tissue.
引用方式:To understand the potential functions of genes with G4, we performed Gene Ontology (GO) term annotation using OmicShare tools (https://www.omicshare.com/tools).
参考文献:
Deng Z, Ren Y, Guo L, et al. Genome-wide analysis of G-quadruplex in Spodoptera frugiperda[J]. International Journal of Biological Macromolecules, 2023, 226: 840-852.
案例4:
发表期刊:International Journal of Biological Macromolecules
影响因子:8.2
发表时间:2023
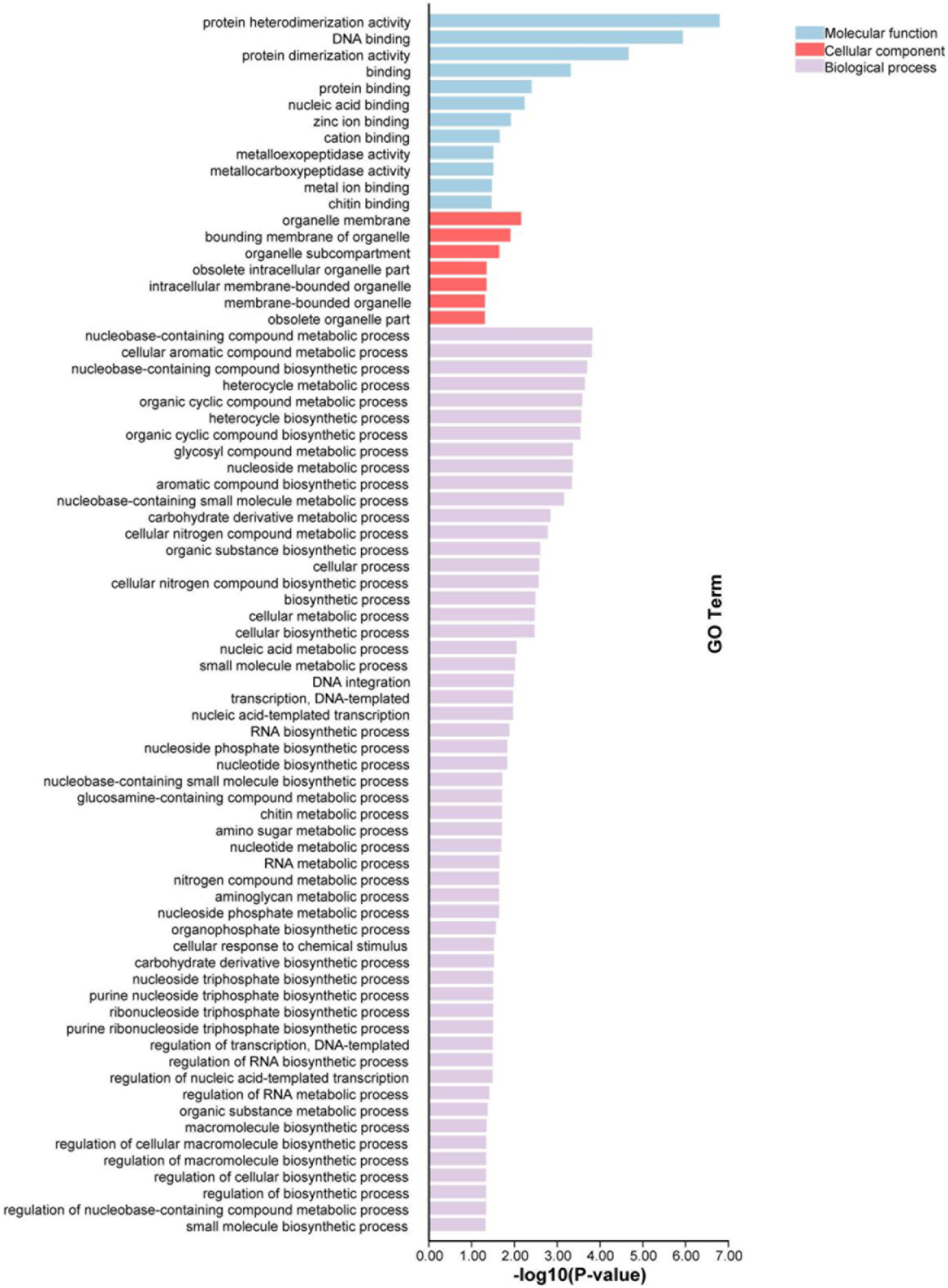
Fig. 4. GO enrichment analysis of G4-rich promoters in S. frugiperda. The genes whose promoters enriched G4 were selected to do enrichment tests in order to reveal the enrichment of gene ontology terms. Statistically significant molecular functions, cellular component and biological process were identified. Each color block represents the gene amount in each tissue.
引用方式:To understand the potential functions of genes with G4, we performed Gene Ontology (GO) term annotation using OmicShare tools (https://www.omicshare.com/tools).
参考文献:
Deng Z, Ren Y, Guo L, et al. Genome-wide analysis of G-quadruplex in Spodoptera frugiperda[J]. International Journal of Biological Macromolecules, 2023, 226: 840-852.
案例5:
发表期刊:Journal of Ethnopharmacology
影响因子:5.4
发表时间:2023
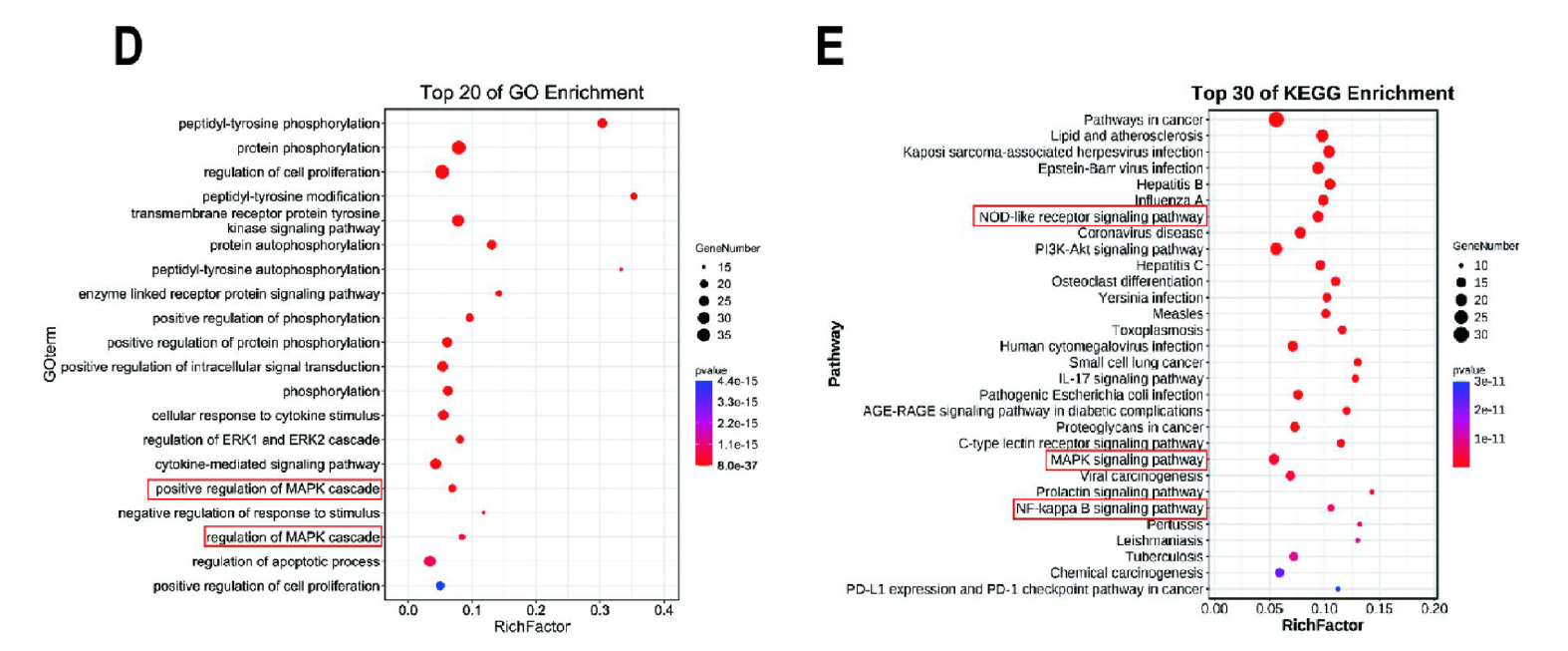
Fig. 2. Network pharmacology predicts the anti-gout mechanism of WWSX. (D) Graph of GO bioprocess enrichment analysis. (E) KEGG mechanism of action enrichment analysis map. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
引用方式:The GO biological process, cellular analysis, molecular function and KEGG pathway bubble map of core targets were constructed on the OmicShare platform, combined with GO and KEGG improvement investigation, the key targets and their biological forms of the organize may well be anticipated.
参考文献:
Bai L, Wu C, Lei S, et al. Potential anti-gout properties of Wuwei Shexiang pills based on network pharmacology and pharmacological verification[J]. Journal of Ethnopharmacology, 2023, 305: 116147.
案例6:
发表期刊:EBioMedicine
影响因子:11.1
发表时间:2022
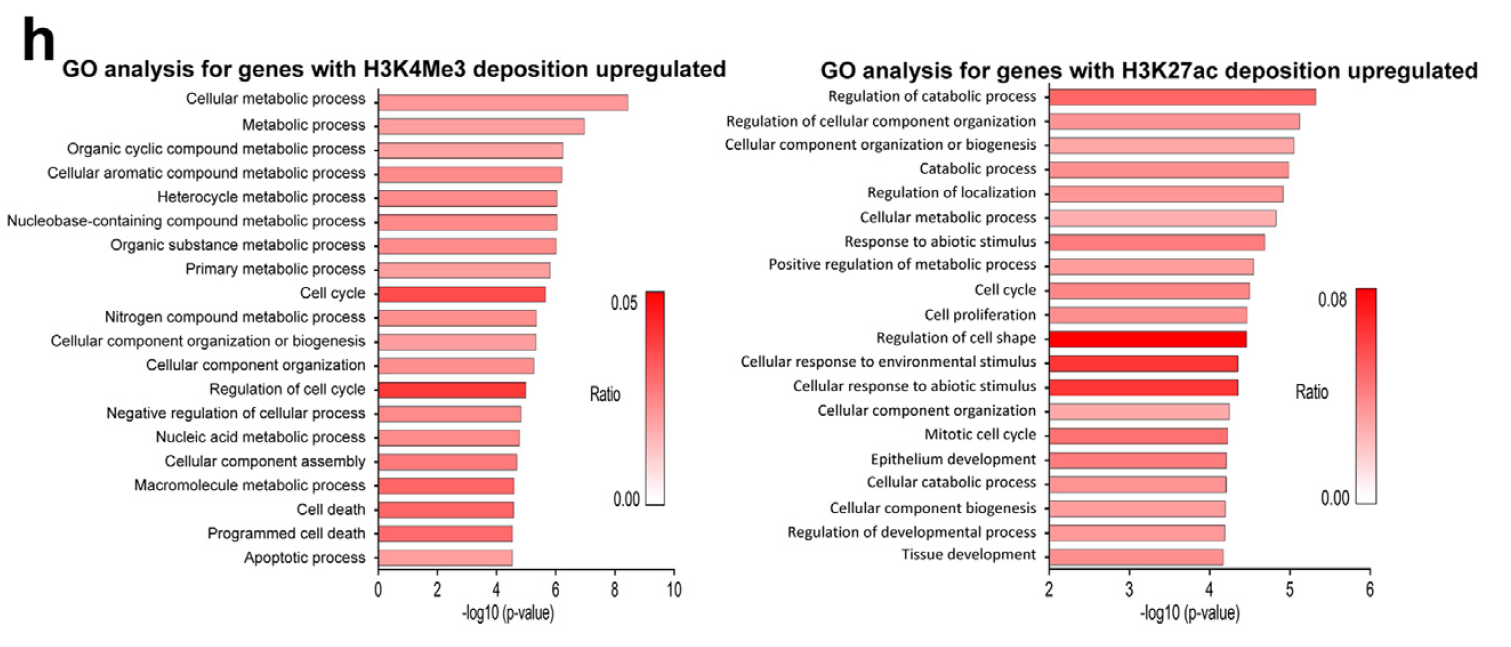
Figure 3. DYRK1A knockdown increases deposition of H3K27ac and H3K4me3 on promoters of cell cycle genes in cardiomyocytes. (h) GO analysis for genes with increased H3K4me3 (left) and H3K27ac (right) deposition on promoter in si-DYRK1A-treated cardiomyocytes.
引用方式:Heatmaps of gene expression, gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway analyses were performed using the OmicShare tools, a free online platform for data analysis (http://www.omicshare.com/tools).
参考文献:
Lan C, Chen C, Qu S, et al. Inhibition of DYRK1A, via histone modification, promotes cardiomyocyte cell cycle activation and cardiac repair after myocardial infarction[J]. EBioMedicine, 2022, 82.
案例7:
发表期刊:International Journal of Molecular Sciences
影响因子:5.6
发表时间:2023
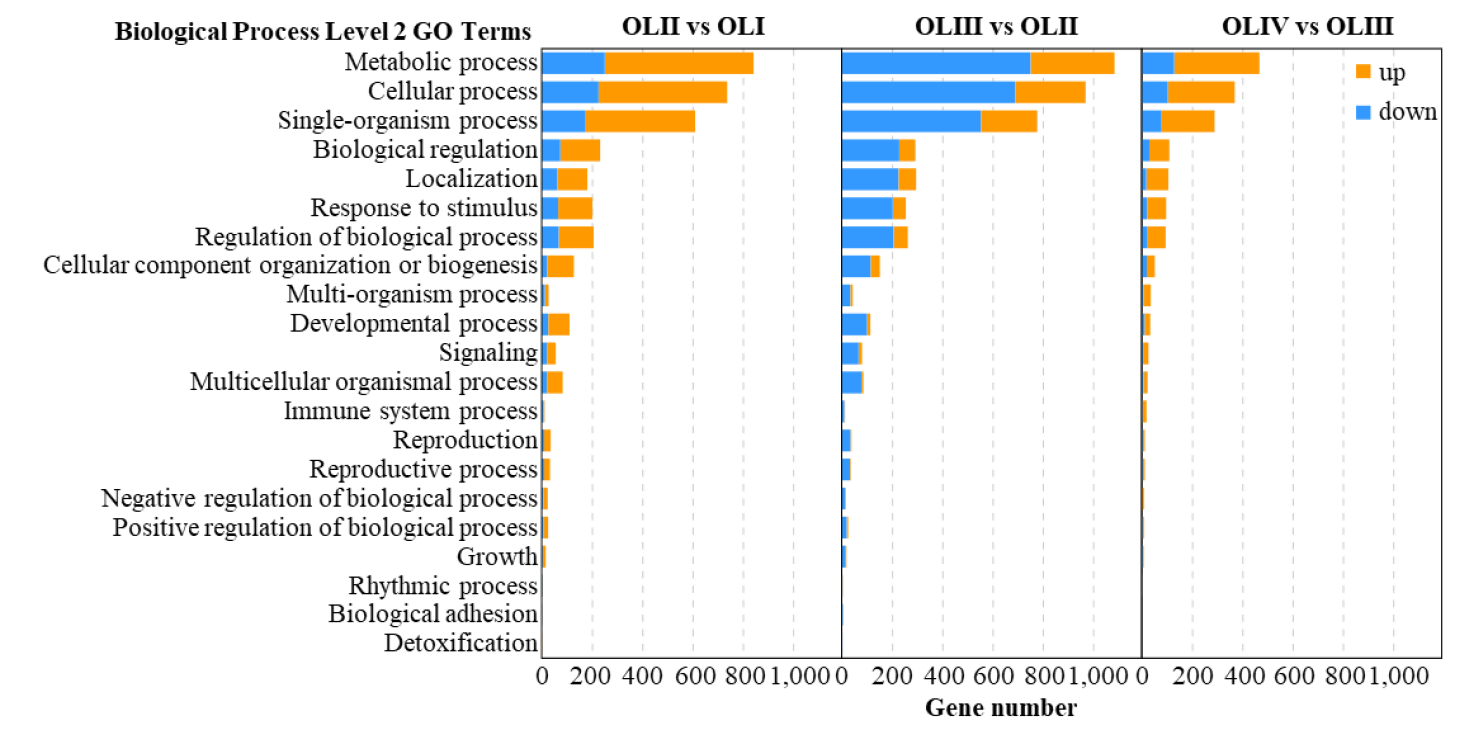
Figure 3. GO functional of the significantly differentially expressed genes of old leaves in diverse pairwise comparisons.
引用方式:GO and KEGG enrichment analyses of DEGs were performed using OmicShare tools in 2021, a free online platform for data analysis (http://www.omicshare.com/tools, 16 August 2022).
参考文献:
Guo H, Zhong Q, Tian F, et al. Transcriptome analysis reveals putative induction of floral initiation by old leaves in tea-oil tree (Camellia oleifera ‘changlin53’)[J]. International Journal of Molecular Sciences, 2022, 23(21): 13021.
案例8:
发表期刊:Precision Medicine Research
发表时间:2022
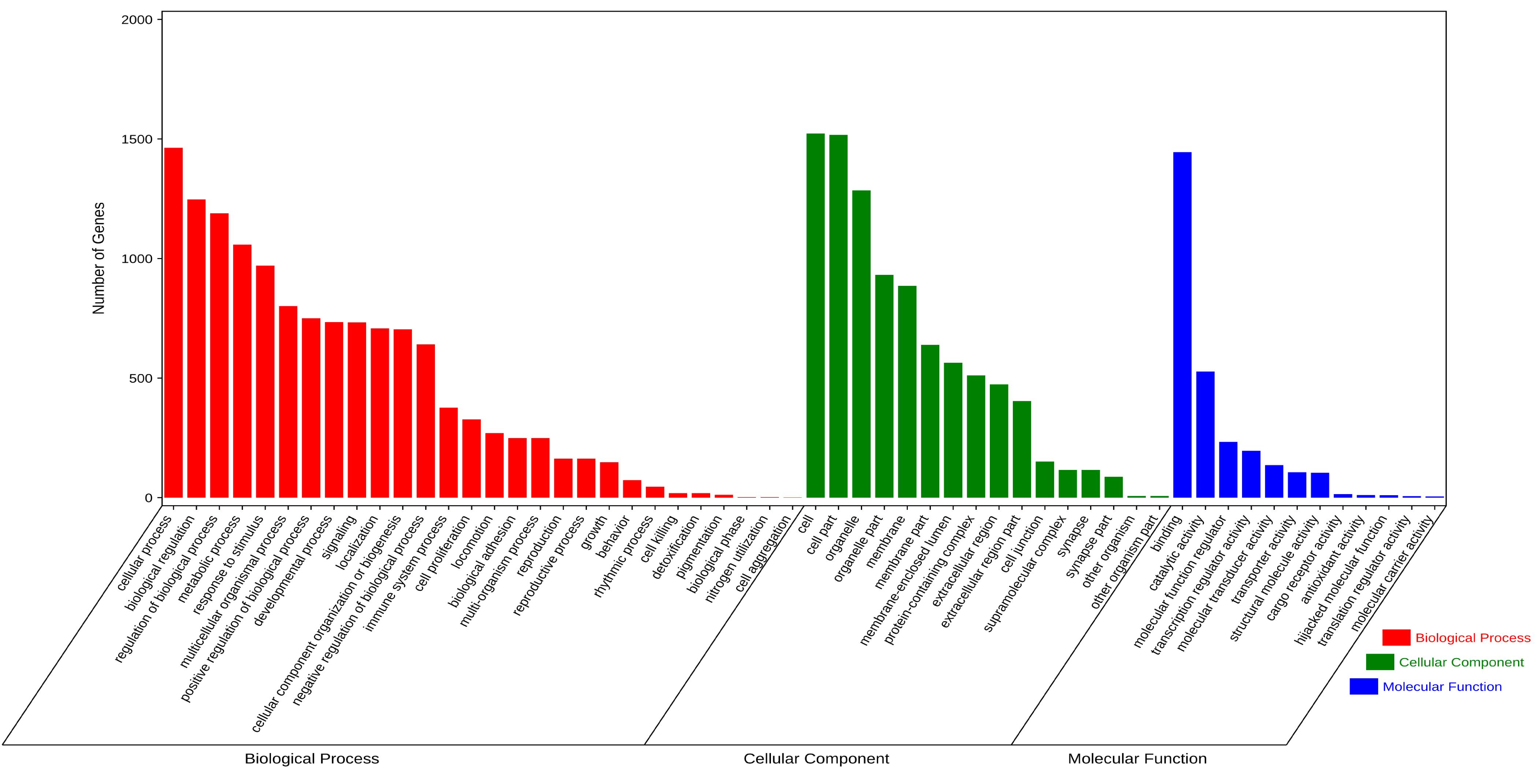
Figure 4. GO analysis of the overlapping DEGs between HCC and BC.
引用方式:The OmicShare database (https://www.omicshare.com/) was used for the visual analysis of KEGG and GO for enrichment analysis. Consequently, we selected the KEGG pathway analysis through the OmicShare database to execute functional annotation on HCC and BC overlapping DEGs.
参考文献:
Xie Z F, Li G G. Identification of overlapping differentially expressed genes in hepatocellular carcinoma, breast cancer, and depression by bioinformatics analysis[J]. Precis Med Res, 2022, 4(3): 11.




 扫码支付更轻松
扫码支付更轻松



 (
(