1. 功能
帮助用户解决常见研究物种的基因ID在不同数据库之间的ID转换,并且提供基因的常见注释信息,为用户研究基因提供方便。
2. 适用物种
拟南芥(Arabidopsis thaliana)、牛(Bos taurus)、山羊(Capra hircus)、秀丽线虫(Caenorhabditis elegans)、果蝇(Drosophila melanogaster)、斑马鱼(danio rerio)、鸡(gallus gallus)、人(homo sapiens)、猕猴(Macaca mulatta)、小鼠(mus musculus)、籼稻(oryza indica)、水稻(oryza sativa)、大鼠(rattus norvegicus)、酵母(Saccharomyces cerevisiae)、番茄(Solanum lycopersicum)、猪(Sus scrofa)、小麦(Triticum aestivum)、玉米(Zea mays)
3. 输入
文件格式:支持txt(制表符分隔)文本文件、csv(逗号分隔)文本文件、以及Excel专用的xlsx格式,同样支持旧版Excel的xls(Excel 97-2003 )格式。
数据格式:一行一个基因ID,以Ensembl Gene ID为例,格式如下:
4. 参数选项
输入文件是否有列名:根据输入文件选择是/否
物种选择:选择输入ID的物种类型
输入ID类型:选择要转换的ID类型,默认可读取4种类型之一,分别是 Gene_stable_ID (Ensembl gene ID) / Gene_name (Gene Symbol) / NCBI_gene_ID (NCBI Entrez Gene ID) / Gene_Synonym
注:4种ID类型的示例文件可点击输入文件旁边的“示例文件”查看
输出ID类型:可选择输出多种ID类型,或者选择输出多种数据库的基因功能注释信息,帮助快速了解该基因功能信息,自行勾选所需即可。
5. 输出文件
xls格式文件
示例文件:小鼠的ensembl ID
1. 参数设置
2. 输出文件
第一列:用户输入的ID,symbol 号
第二列:小鼠的ensembl ID
第三列:NCBI ID号,通常是数值
第四列:基因同义名
第五列:基因的注释信息,如基因功能,表达某蛋白
第六列:KEGG A分类,一般有7大类
第七列:KEGG B分类,比A分类的信息更详细
第八列:通路ID,即ko号,还有通路详细名称,如TGFβ信号通路
第九列:K号,可用与KEGG富集分析
第十列:GO注释细胞组分
第十一列:GO注释分子功能
第十二列:GO注释生物学途径
Q1. 为什么输出没有结果,如下图?
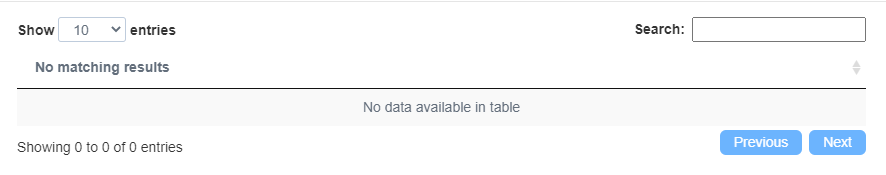
没有输出结果可能原因:
①物种选错了;
②上传的文件中,基因ID类型跟参数选择不对应。
Q2. 为什么输出结果,数目比我提交时多?
比如 一个NCBI_gene_ID可能对应多个Gene Synonym(如:3804对应Gene Synonym有CD158B2、p58、NKAT2),工具会把所有结果输出。
Q3:为什么有部分基因没有结果?
如果有部分基因没有数据,有可能是该基因类型选错了。
比如下图中"CD89""103AS"类型是Gene_Synonym,在分析时选了”Gene_name“则比对不上,无结果了。

Q4. 提交时报错常见问题:
1.提交时显示X行X列空行/无数据,请先自查表格中是否存在空格或空行,需要删掉。
2.提交时显示列数只有1列,但表格数据不止1列:列间需要用分隔符隔开,先行检查文件是否用了分隔符。
其它提示报错,请先自行根据提示修改;如果仍然无法提交,可通过左侧导航栏的“联系客服”选项咨询OmicShare客服。
Q3. 提交的任务完成后却不出图该怎么办?
主要原因是上传的数据文件存在特殊符号所致。可参考以下建议逐一排查出错原因:
(1)数据中含中文字符,把中文改成英文;
(2) 数据中含特殊符号,例如 %、NA、+、-、()、空格、科学计数、罗马字母等,去掉特殊符号,将空值用数字“0”替换;
(3)检查数据中是否有空列、空行、重复的行、重复的列,特别是行名(一般为gene id)、列名(一般为样本名)出现重复值,如果有删掉。
排查完之后,重新上传数据、提交任务。如果仍然不出图,可通过左侧导航栏的“联系客服”选项咨询OmicShare客服。
引用OmicShare Tools的参考文献为:
Mu, Hongyan, Jianzhou Chen, Wenjie Huang, Gui Huang, Meiying Deng, Shimiao Hong, Peng Ai, Chuan Gao, and Huangkai Zhou. 2024. “OmicShare tools: a Zero‐Code Interactive Online Platform for Biological Data Analysis and Visualization.” iMeta e228. https://doi.org/10.1002/imt2.228案例1:
发表期刊:Journal of Hainan Medical College
发表时间:2020
引用方式:
“to obtain the Gene ID of the target, and then convert it into the Ensembl Gene ID through the Ensembl genome database, and import it into the OmicShare cloud platform“
参考文献:
Zhou M Q, Yang L P, Ma H J, et al. Network pharmacological study of Qingfei Paidu Decoction intervening on cytokine storm mechanism of COVID-19[J]. Journal of Hainan Medical College, 2020, 26(10).




 扫码支付更轻松
扫码支付更轻松


