1.功能:
使用富集分析结果表格绘制富集圈图。
2.输入文件:
输入矩阵表格文件,其中:
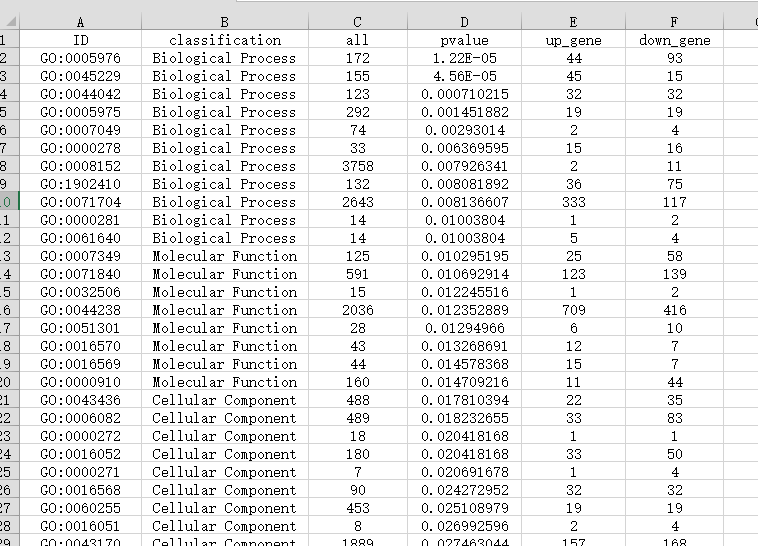
第1列:ID,例如输入GO term的ID信息(注意:第一列不能出现重复ID)。
第2列:Classification,该ID对应的分类名称,图形中的功能图例根据该列信息进行分类展示。
第3列:all,表示该样本在这个分类中鉴定到的基因总数,即背景基因。
第4列:P值/Q值,基于P值或Q值做图,只输入其一。
第5列、第6列:有两种格式可选择输入,当区分上下调基因时,则第5列、第6列分别输入上调、下调基因的数量。当不区分上下调时,则仅需输入第5列的差异基因总数。
示例文件一(共6列,区分上下调基因的数据表格):
示例文件二(共计5列,不区分上下调基因)
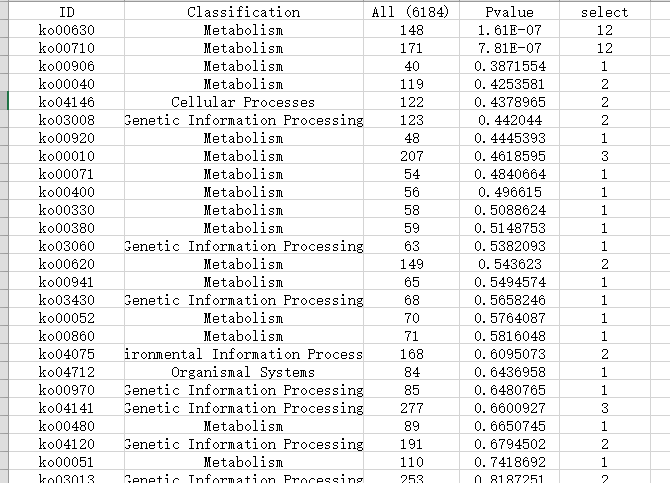
(备注:最终绘制的图中,富集值(RichFactor)=(5列+6列)/3列)
输入文件类型:输入的文件必须是带有表头的以tab分隔的.txt格式文件。表格名称以字母、数字、下划线组成,不可以有后缀名称。可以选择在excel中将数据打开,然后另存为“文本文件(制表符分隔)(*.txt)”
3.参数选择:
a.参数:选择绘图使用的p值或是q值(该信息将展示在图例中)
b.用于作图的行数:下拉选择用于作图的数据行数,推荐使用不超过25行数据作图,绘图行数太多可能导致文字重叠,影响观感。
3.图形调整参数
全局修改:
a.字体/名称大小:调整对应位置的字体大小
b.基础图例:勾选基础图例展示位置
c.勾选展示:功能图例、显著性图例、功能名称
d.自适应:圈外刻度自适应/输入刻度范围、圈内柱形图自适应/输入值范围
基础配色:
a.名称颜色:修改第一圈ID文字颜色
b.上下调基因配色:修改上下调基因配色
c.功能配色:可调整各功能配色
区间配色:
可调整显著性数值(-log10(P value)/-log10(Q value))区间数值,并可对每个子区间进行配色选择。
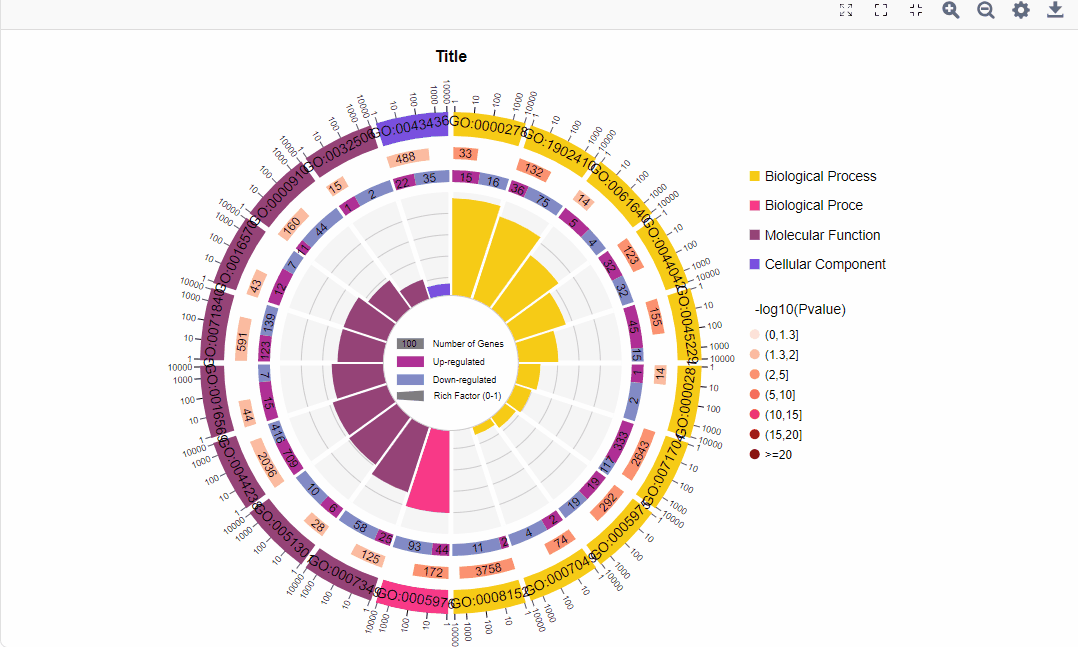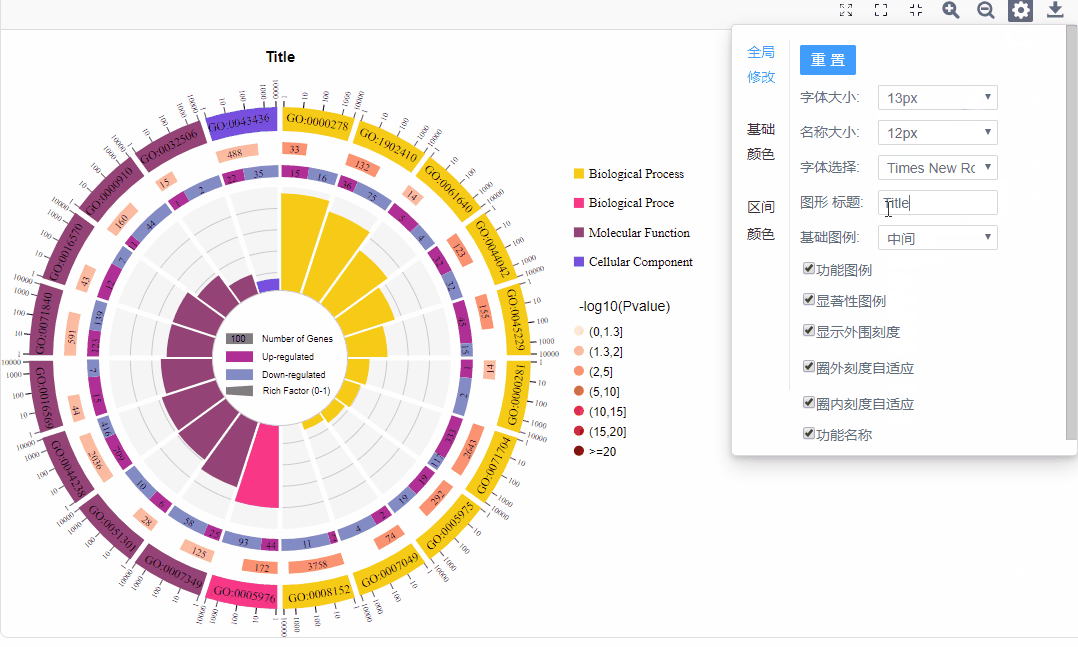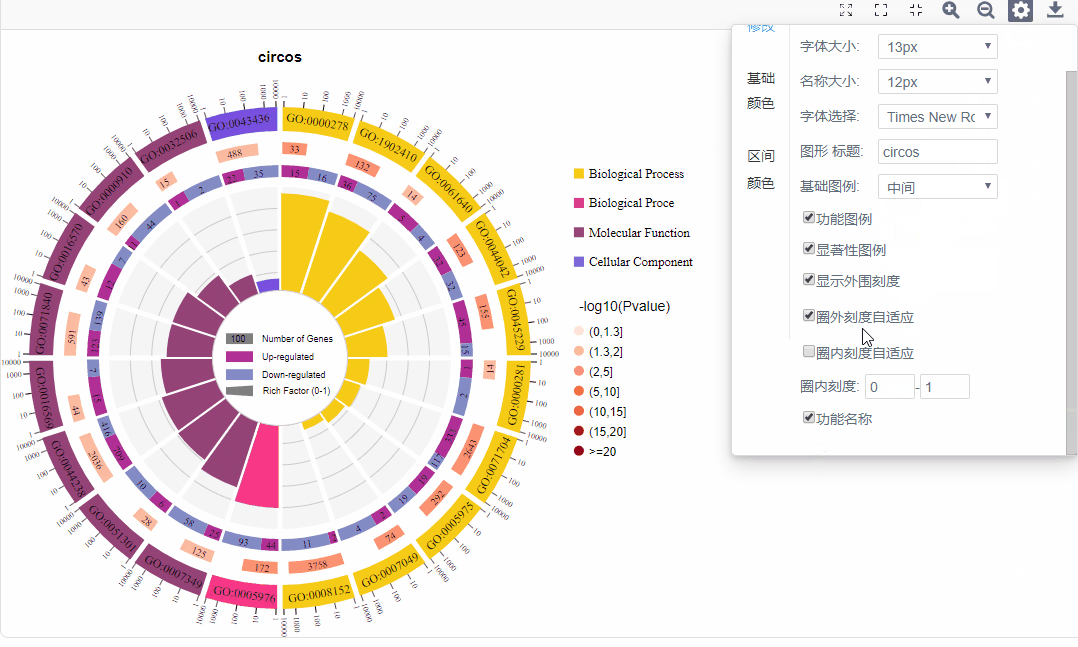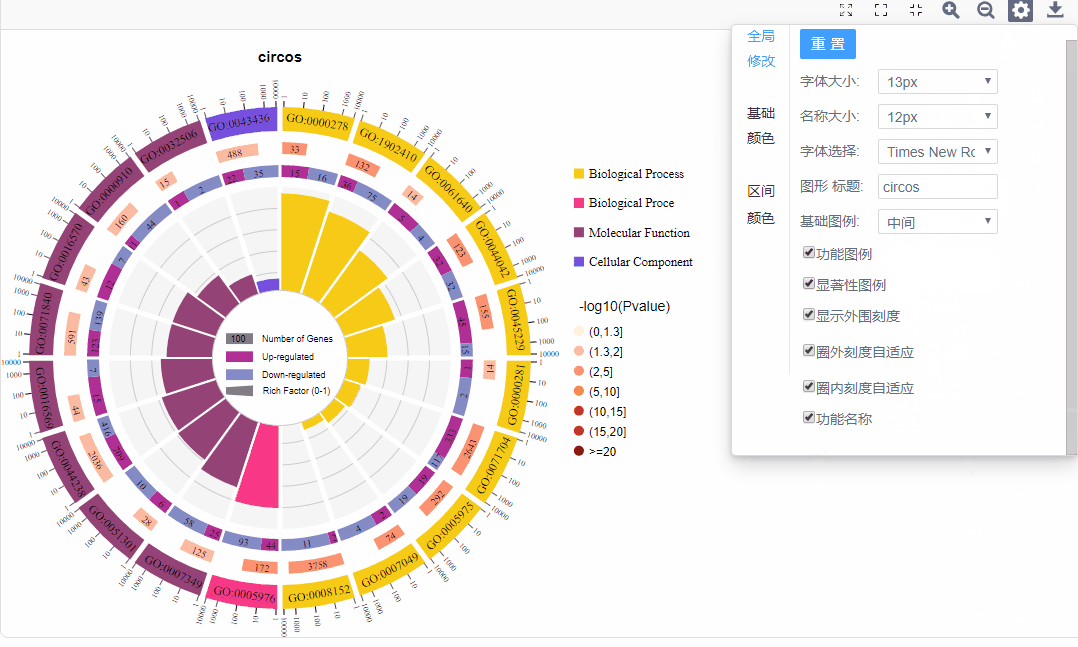
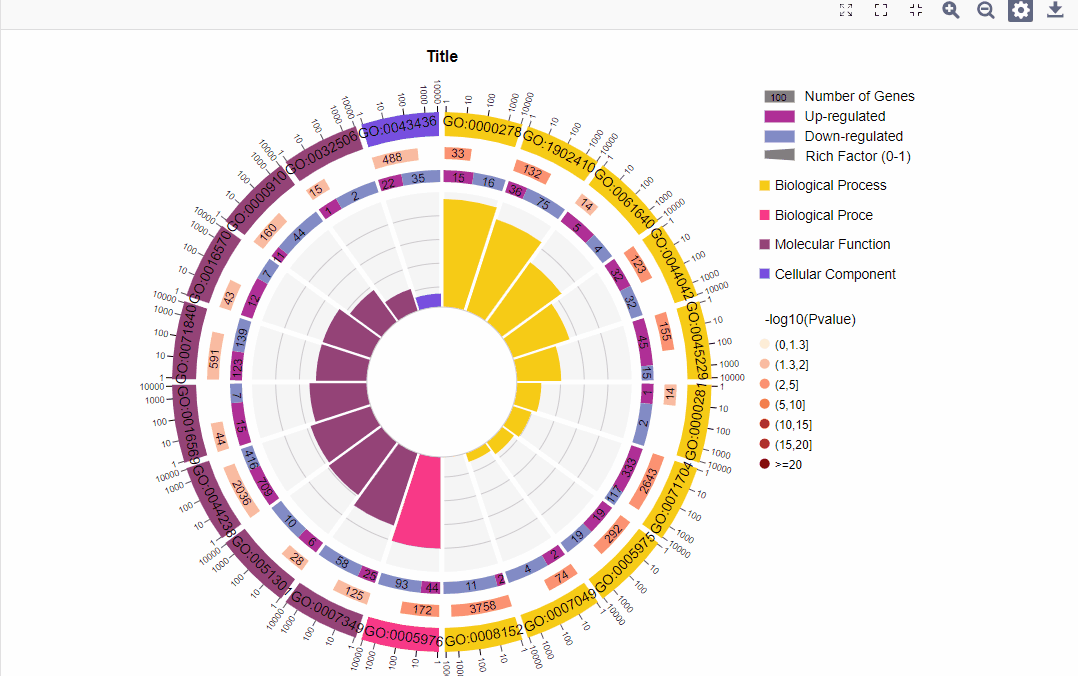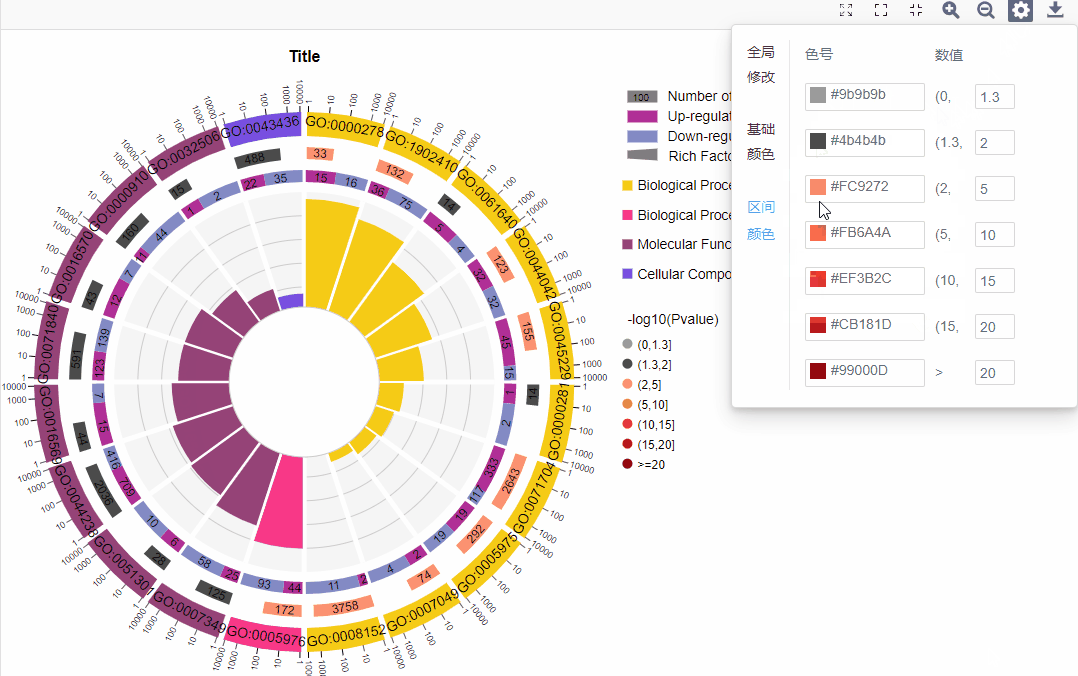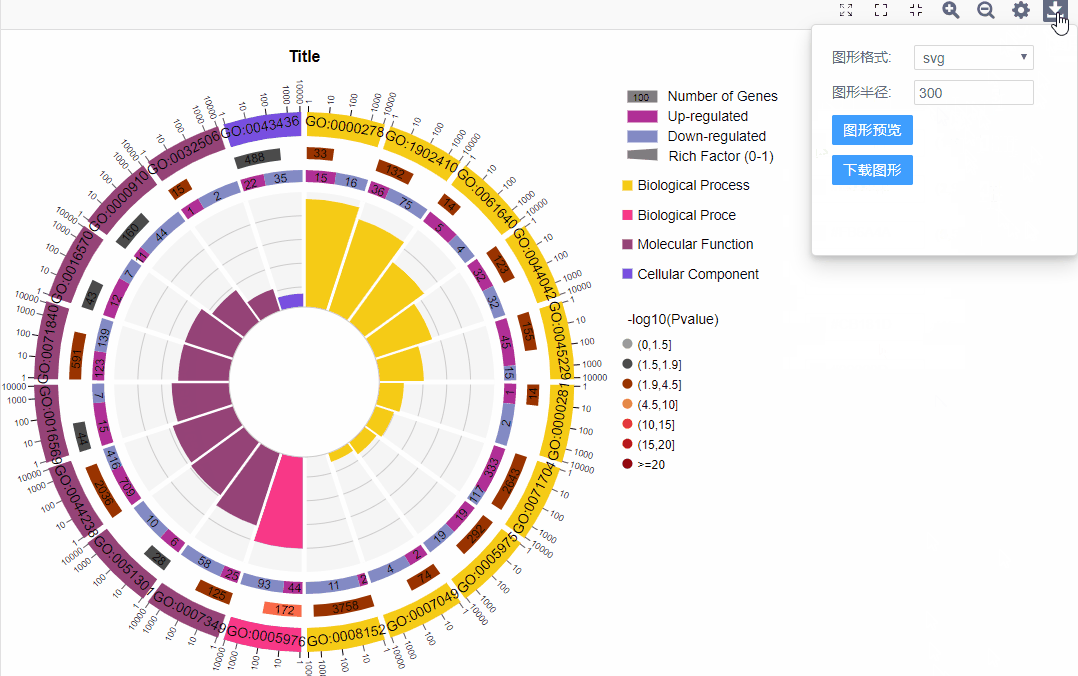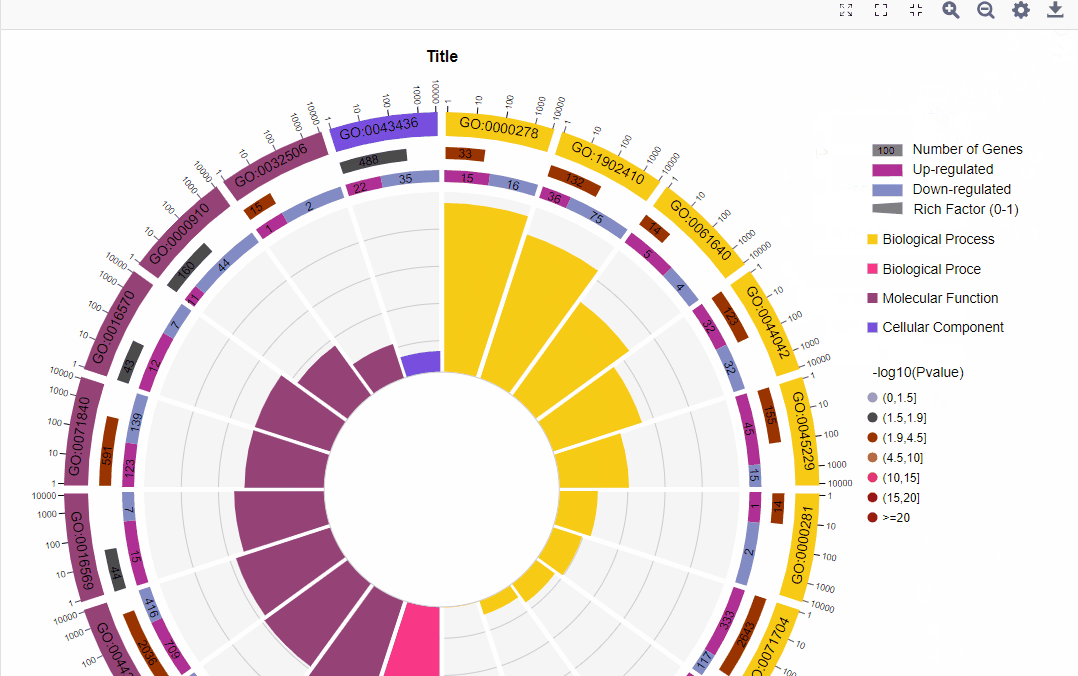
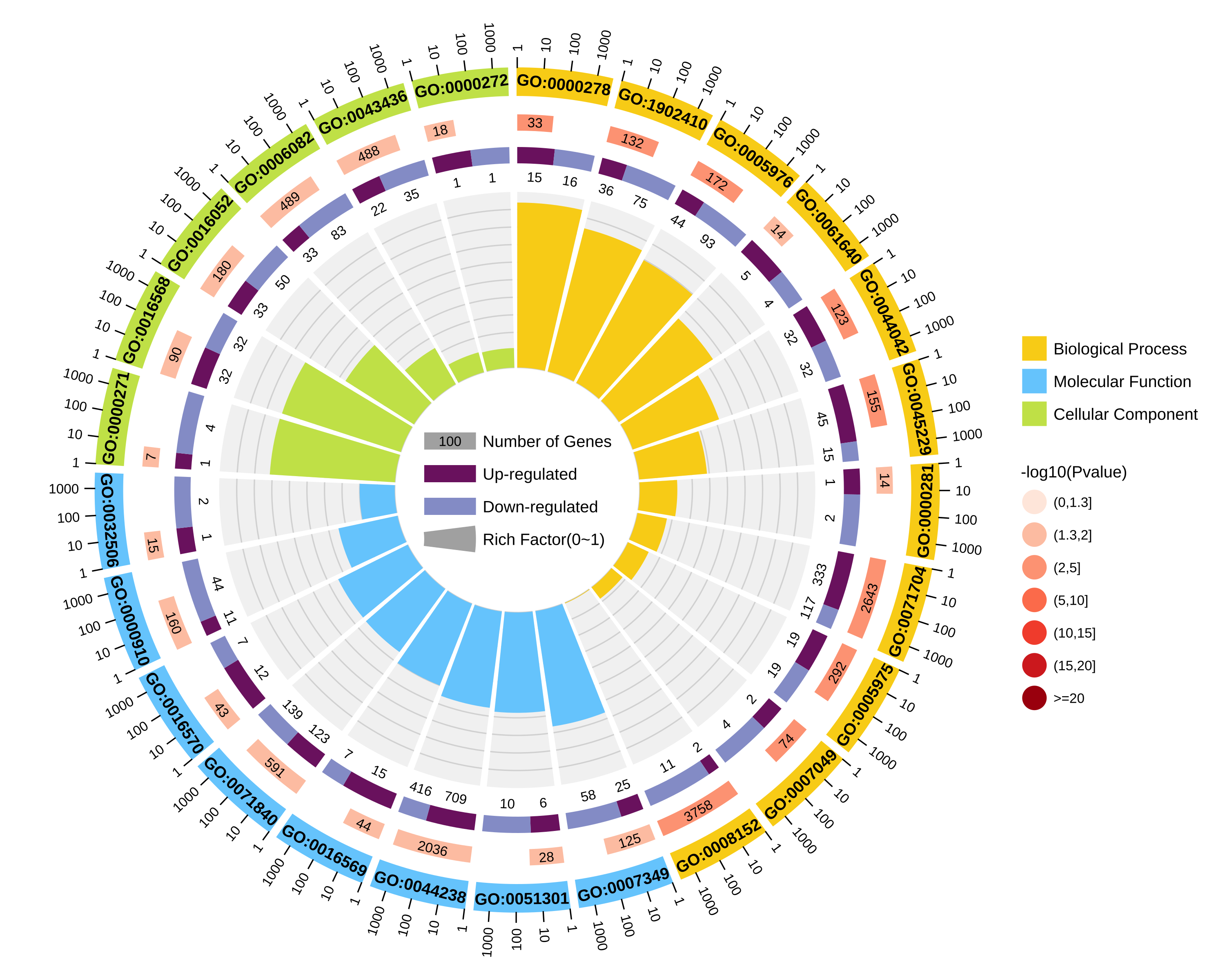
结果图示例如下:
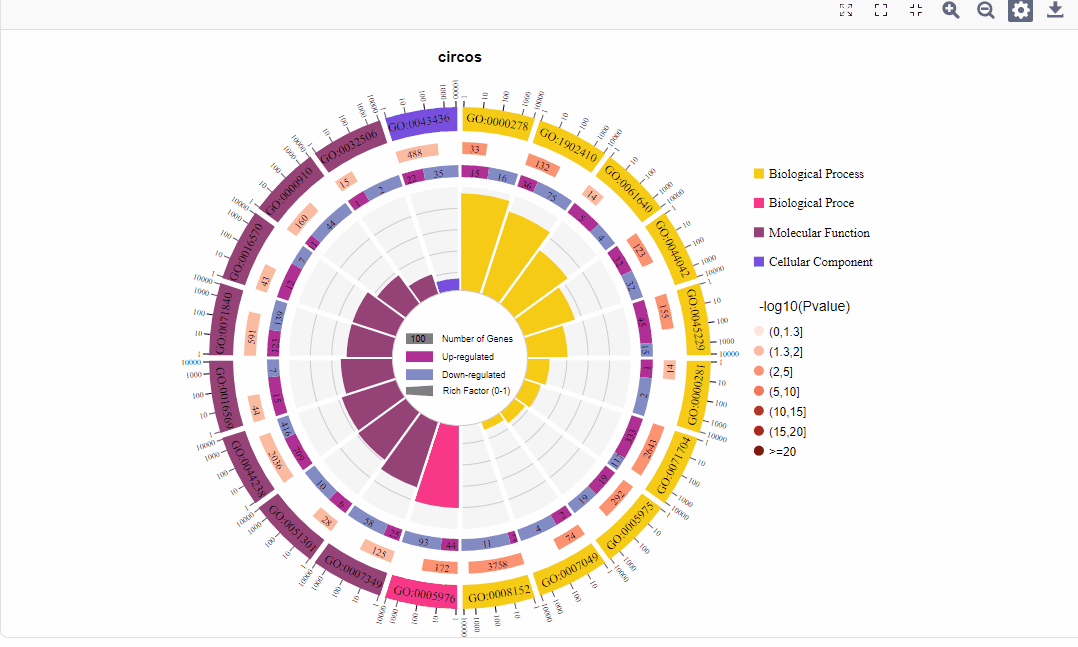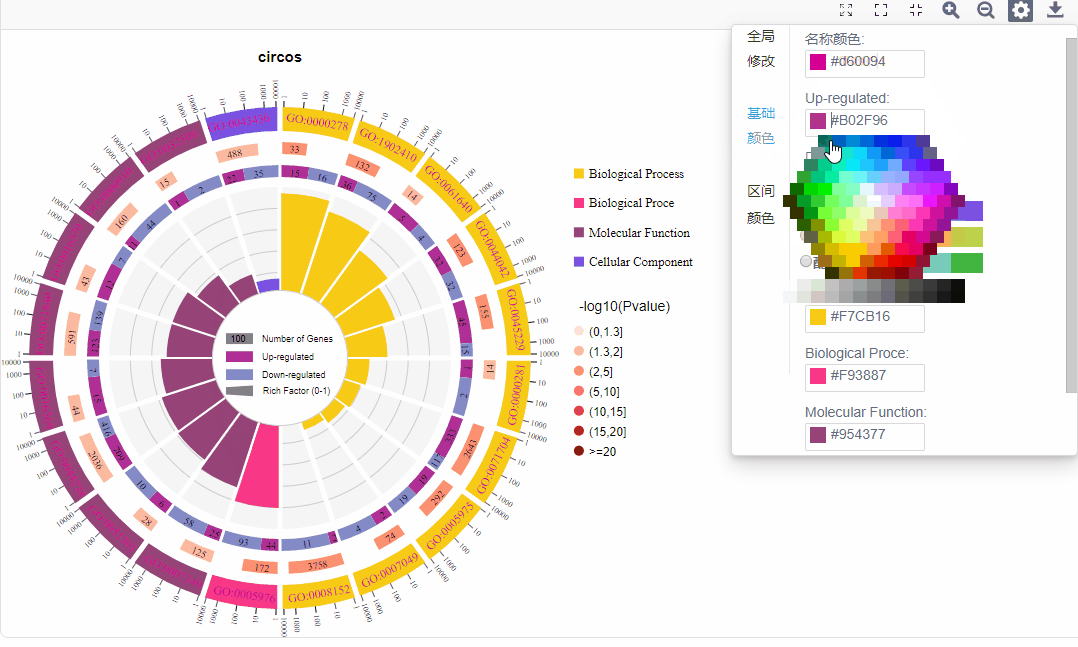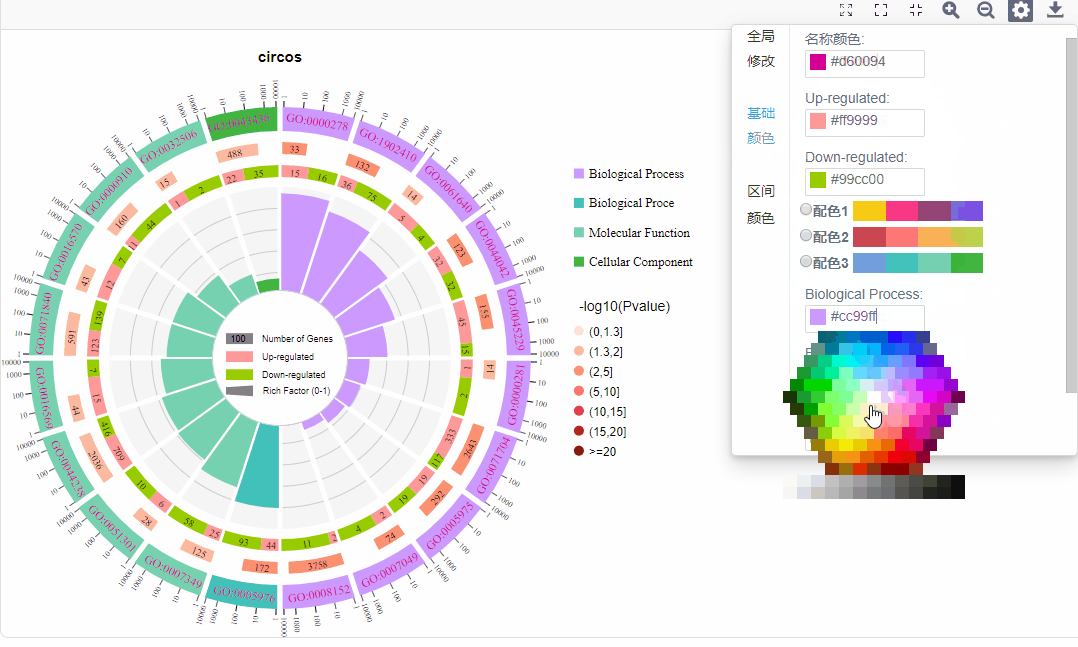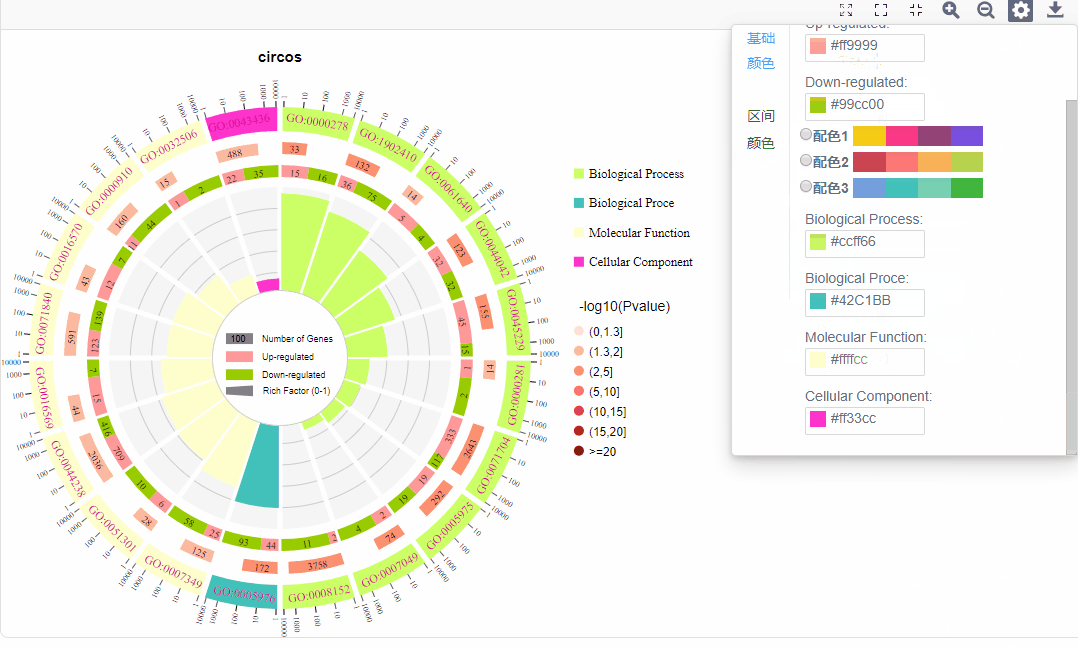
1.区分上下调基因图形:

2.不区分上下调基因图形:
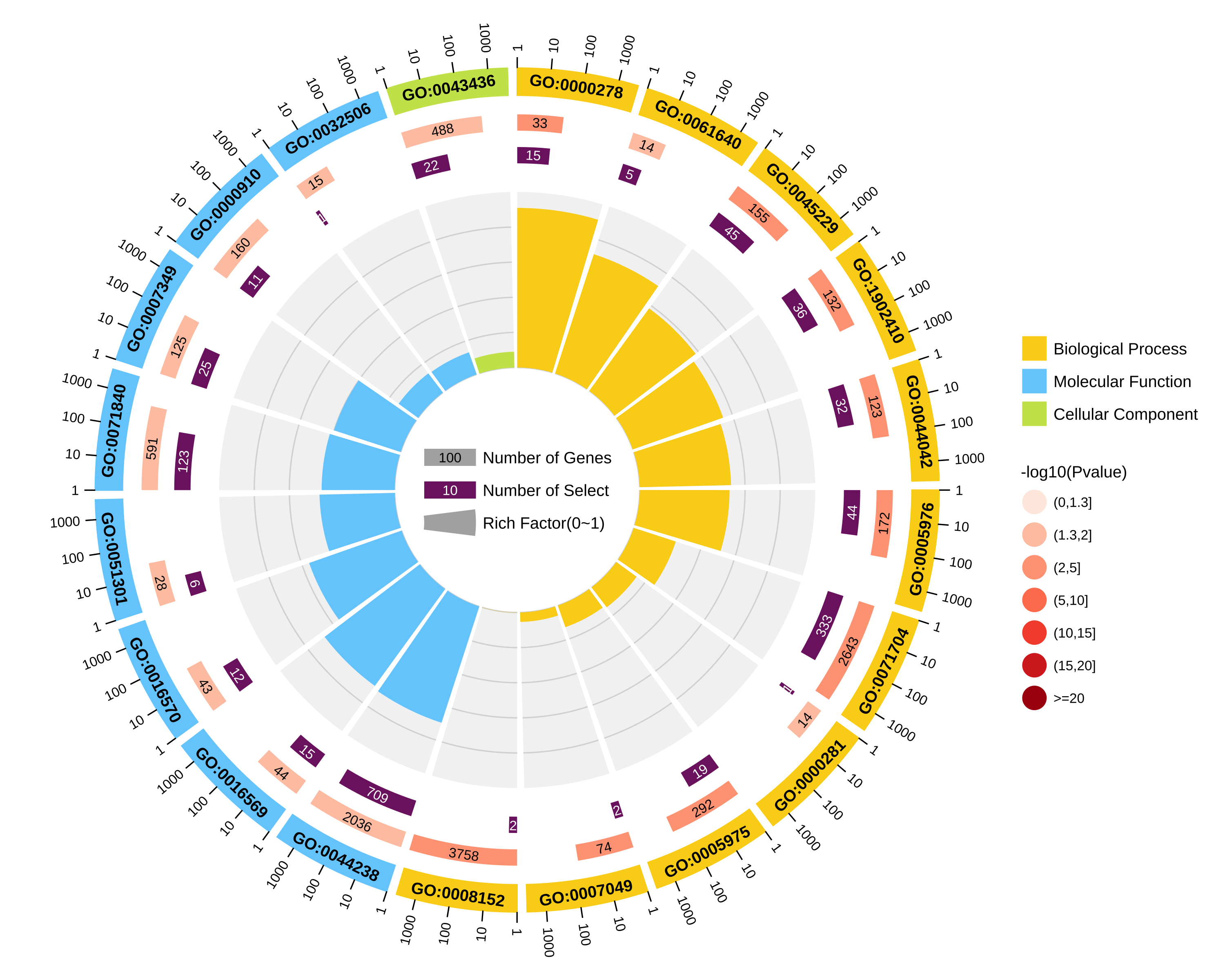
图形解读:
|
模块功能 |
使用富集圈图展示富集分析的结果。 |
|
图形解读 |
从外到内共四圈。 1.第一圈:富集的分类,圈外为基因数目的坐标尺。不同的颜色代表不同的分类; 2.第二圈:背景基因中该分类的数目以及Q值或P值。基因越多条形越长,值越小颜色越红; 3.第三圈:上下调基因比例条形图,深紫色代表上调基因比例,浅紫色代表下调基因比例;下方显示具体的数值;当输入的差异基因数量只有一列(未区分上下调)时,第三圈显示前景基因的总数目; 4.第四圈:各分类的RichFactor值(该分类中前景基因数量除以背景基因数量),背景辅助线每个小格表示0.1。 |
Q1. 上传的数据需要保存成什么格式?文件名称和拓展名有没有要求?
OmicShare当前支持txt(制表符分隔)文本文件、csv(逗号分隔)文本文件、以及Excel专用的xlsx格式,同样支持旧版Excel的xls(Excel 97-2003 )格式。如果是核酸、蛋白序列文件,必须为FASTA格式(本质是文本文件)。
文件名可由英文和数字构成,文件拓展名没有限制,可以是“.txt”、“.xlsx”、“.xls”、“.csv”“.fasta”等,例如 mydata01.txt,gene02.xlsx 。
Q2. 提交时报错常见问题:
1.提交时显示X行X列空行/无数据,请先自查表格中是否存在空格或空行,需要删掉。
2.提交时显示列数只有1列,但表格数据不止1列:列间需要用分隔符隔开,先行检查文件是否用了分隔符。
其它提示报错,请先自行根据提示修改;如果仍然无法提交,可通过左侧导航栏的“联系客服”选项咨询OmicShare客服。
Q3. 提交的任务完成后却不出图该怎么办?
主要原因是上传的数据文件存在特殊符号所致。可参考以下建议逐一排查出错原因:
(1)数据中含中文字符,把中文改成英文;
(2) 数据中含特殊符号,例如 %、NA、+、-、()、空格、科学计数、罗马字母等,去掉特殊符号,将空值用数字“0”替换;
(3)检查数据中是否有空列、空行、重复的行、重复的列,特别是行名(一般为gene id)、列名(一般为样本名)出现重复值,如果有删掉。
排查完之后,重新上传数据、提交任务。如果仍然不出图,可通过左侧导航栏的“联系客服”选项咨询OmicShare客服。
Q4.下载的结果文件用什么软件打开?
OmicShare云平台的结果文件(例如,下图为KEGG富集分析的结果文件)包括两种类型:图片文件和文本文件。
图片文件:
为了便于用户对图片进行后期编辑,OmicShare同时提供位图(png)和矢量图(pdf、svg)两种类型的图片。对于矢量图,最常见的是pdf和svg格式,常用Ai(Adobe illustrator)等进行编辑。其中,svg格式的图片可用网页浏览器打开,也可直接在word、ppt中使用。
文本文件:
文本文件的拓展名主要有4种类型:“.os”、“.xls”、“.log”和“.txt”。这些文件本质上都是制表符分隔的文本文件,使用记事本、Notepad++、EditPlus、Excel等文本编辑器直接打开即可。结果文件中,拓展名为“.os”文件为上传的原始数据;“.xls”文件一般为分析生成的数据表格;“.log”文件为任务运行日志文件,便于检查任务出错原因。
Q5. 提交的任务一直在排队怎么办?
提交任务后都需要排队,1分钟后,点击“任务状态刷新”按钮即可。除了可能需运行数天的注释工具,一般工具数十秒即可出结果,如果超出30分钟仍无结果,请联系OS客服,发送任务编号给OmicShare客服,会有专人为你处理任务问题。
Q6. 结果页面窗口有问题,图表加载不出来怎么办?
尝试用谷歌浏览器登录OmicShare查看结果文件,部分浏览器可能不兼容。
引用OmicShare Tools的参考文献为:
Mu, Hongyan, Jianzhou Chen, Wenjie Huang, Gui Huang, Meiying Deng, Shimiao Hong, Peng Ai, Chuan Gao, and Huangkai Zhou. 2024. “OmicShare tools: a Zero‐Code Interactive Online Platform for Biological Data Analysis and Visualization.” iMeta e228. https://doi.org/10.1002/imt2.228案例1:
发表期刊:Signal Transduction and Targeted Therapy
影响因子:39.3
发表时间:2022
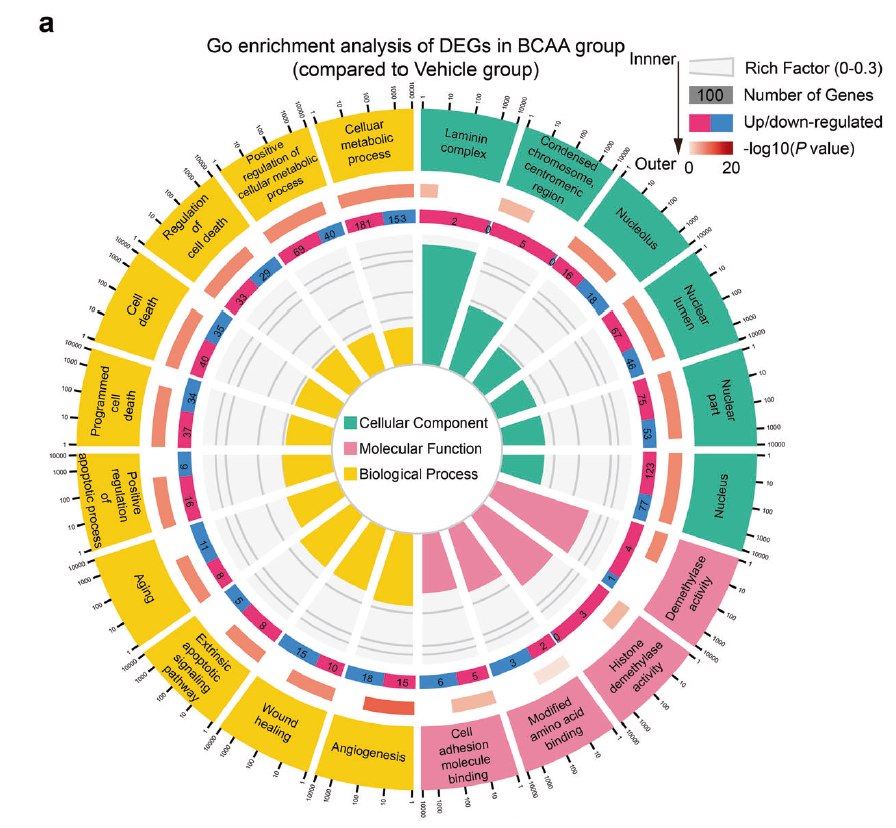
Fig. 3 BCAA accelerated H3K9me3 loss in ADSCs upon exposure to detrimental stress. a GO enrichment of the DEGs in ADSCs treated with vehicle or BCAA (3.432 mM) under hydrogen peroxide (100 μM) stress as identified by RNA-seq. Twenty significantly enriched GO terms are shown.
引用方式:
Gene Ontology (GO) enrichment analysis was performed and the results visualized with the
OmicShare tool, an online platform for data analysis (https://www.omicshare.com/tools/Home/Soft/enrich_circle).
参考文献:
Zhang F, Hu G, Chen X, et al. Excessive branched-chain amino acid accumulation restricts mesenchymal stem cell-based therapy efficacy in myocardial infarction[J]. Signal Transduction and Targeted Therapy, 2022, 7(1): 171-171.
案例2:
发表期刊:International Journal of Molecular Sciences
影响因子:5.6
发表时间:2022

Figure 4. Gene co-expression network analysis. (c) KO (KEGG Ontology) enrichment circle diagram of cyan module (from the outside to the inside, the first circle represents the top 20 enrichment pathways, and the number outside the circle is the coordinate ruler of the number of genes; The second circle represents the number and Q value of background genes in this pathway, and the more genes, the longer the bar; The third circle represents the number of the DEGs in this pathway; The fourth circle represents the value of Rich Factor in each pathway)
引用方式:By means of the OmicShare tools (https://www.omicsmart.com/, accessed on 1 November 2021), a weighted gene co-expression network analysis was constructed to explore molecular regulatory mechanisms involved in photosynthesis.
参考文献:
Zhang L, Zhang Z, Fang S, et al. Metabolome and transcriptome analyses unravel the molecular regulatory mechanisms involved in photosynthesis of Cyclocarya paliurus under salt stress[J]. International Journal of Molecular Sciences, 2022, 23(3): 1161.
案例3:
发表期刊:Pharmacological Research
影响因子:9.3
发表时间:2023
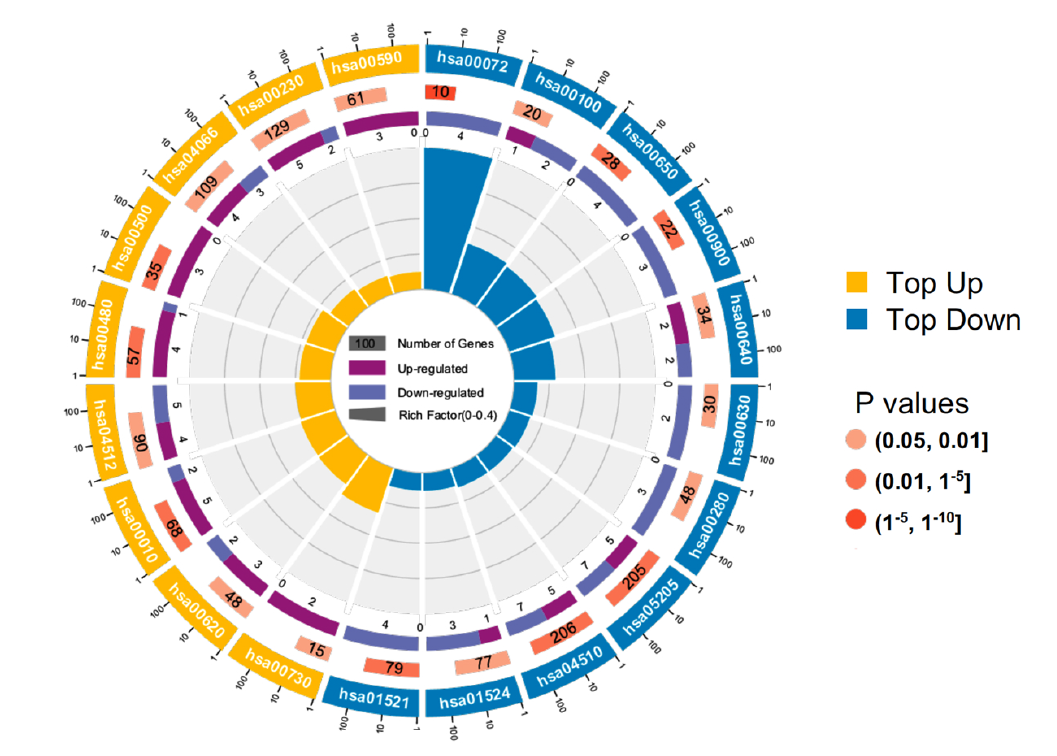
Fig. 4. Quantitative proteomics analysis of PC tumor tissues revealed the changed metabolic enzymes of AMP. (A) The KEGG pathway enrichment analysis based on differential proteins of PC tumor tissues.
引用方式:KEGG enrichment pathways with P-value less than 0.05 and relevant to this study were characterized using the OmicShare tools (https://www.omicshare.com/tools).
参考文献:
Liu J, Jing W, Wang T, et al. Functional metabolomics revealed the dual-activation of cAMP-AMP axis is a novel therapeutic target of pancreatic cancer[J]. Pharmacological Research, 2023, 187: 106554.




 扫码支付更轻松
扫码支付更轻松