输入示例文件:
文件一:sample_table1
文件二:sample_table2
说明:示例数据是用富集到该通路的基因数渐变节点大小,用富集分析后的检验值FDR来渐变节点颜色。您可以选择其他数据定义节点属性。
根据示例文件格式整理好数据,上传,提交即可。
在结果展示中可根据自己的需求修改网络图。
输入动态演示:(输入一次,可以得到不同类型的网络图)
1. 正常网络图:
a) 全局修改可调整布局,字体,字号,是否选用标签及标签的位置
b) 节点修改可调整形状,透明度,颜色,大小
c) 节点边框可调整边框粗细,颜色
d) 线的修改可调整线的类型,颜色,粗细
e) 目标点线可自定义点线的形状,颜色,大小等,也可删除节点等,主要用于点缀重要的点和线。

最后保存的结果:

2. 补充连接通路的网络图:
在全局修改点击补充连接通路
其他设置同正常网络图

最后保存的结果:

3. 补充相邻通路的网络图:
在全局修改点击补充所有相邻通路
其他设置同正常网络图

最后保存的结果:

1. 功能:
网络图是生物信息学中常用的显示不同节点之间关联方向与关联程度的可视化方法。在富集分析中,网络图常被用于表示富集路径之间的相互作用关系,发现上下游信号通路,寻找核心通路,或者补充中断的通路,使用户从全局了解网络通路的关系,并且展示通路与基因的关系。在生物体内,不同基因相互协调行使其生物学功能,基于Pathway的分析有助于更进一步了解基因的生物学功能。
2. 应用范围:
适用于KEGG富集分析后,用网络图来呈现各个通路之间的关系,并且还将通路与基因的关系呈现出来,使用户从全局了解通路与通路、通路与基因的关系。
3. 输入:(格式如下,支持txt(制表符分隔)文本文件、csv(逗号分隔)文本文件、以及Excel专用的xlsx格式,同样支持旧版Excel的xls(Excel 97-2003 )格式。)
(1)通路文件(必需):展示各个通路及属性
第一列为kegg的map ID,即ko号
第二列属性用来渐变节点大小,第三列属性用来渐变节点颜色,其中数据的类型没有固定,您可以选择各种数据来渐变节点大小,只需将数据放在相应列即可。或者直接输入通路ko号,不添加节点属性也行。(注意:属性文件只能输入数值,不支持文本类)。
下表的示例中,用富集到该通路的基因数目来渐变节点大小,用富集分析后的检验值FDR取 -lg(FDR) 值来渐变节点颜色。如果您想用其他数据作为节点属性也行。
|
Map_ID |
Gene_number |
FDR |
|
ko01001 |
2 |
0.00001 |
|
ko04812 |
4 |
0.000003 |
|
ko03013 |
12 |
1e-10 |
|
…… |
…… |
…… |
注意:输入格式可以为3列,也可以只输入第1列。
提示:建议输入10个通路左右,不然图形中的节点太多,影响图形美观;可以根据实验设计想要揭示的问题来选择与实验现象相关的通路或者重点关注的显著富集通路。
(2)通路ko号与基因ID关系文件(非必需):用于在网络图中展示通路与基因的关系
第一列为想展示的基因所在的通路ko号
第二列为基因ID。可以自定义,或者使用symbol号。
|
ko number |
gene_ID (or symbol) |
|
ko04014 |
gene1 |
|
ko04014 |
gene2 |
|
ko04921 |
gene3 |
|
…… |
…… |
建议:输入的基因节点也不要太多,不然影响整体布局。可以选择展示经过实验验证的重要基因,或查阅文献后得知的重要基因。
4. 参数设置:
(1)补充连接通路:选择”是/否“。在网络图中显示可以将输入的两个kegg通路连在一起的通路(这类通路不在输入的通路列表里,而是由KEGG连接通路数据库中补充得出,补充的通路是中间的,使得输入的孤立通路连接形成完整网络)。这类通路节点颜色默认为灰色,连接线用虚线连接。
(2)补充所有相邻通路:选择”是/否“。在网路图中显示与输入的KEGG通路相邻的所有补充通路。(这类通路不在输入的通路列表里,而是由KEGG连接通路数据库中得出,补充相邻通路,使得网络信息更加完整)。这类通路节点颜色默认为灰色,连接线用虚线连接。
(3)其他参数设置:如节点大小,颜色,性状,连接线的颜色,类型等可以根据需求设置。
5. 输出:
(1)节点文件:(Nodes)
|
ID |
name |
Type |
Gene number |
Degree |
FDR |
|
ko04014 |
Ras signaling pathway |
Pathway |
41 |
2 |
8.56E-83 |
|
ko04024 |
cAMP signaling pathway |
pathway |
16 |
6 |
3.42E-25 |
|
… |
… |
… |
… |
… |
… |
|
ko23120 |
… |
Add_link |
- |
- |
- |
|
ko00211 |
… |
Add_link |
- |
- |
- |
|
…… |
…… |
…… |
…… |
…… |
…… |
|
ko00101 |
…… |
Add_Adjacent |
- |
- |
- |
|
ko00112 |
…… |
Add_Adjacent |
- |
- |
- |
|
…… |
…… |
…… |
…… |
…… |
…… |
|
- |
Unigene0003256(Rasa4) |
gene |
- |
- |
- |
|
- |
Unigene0003714(B0563.7) |
Gene |
- |
- |
- |
|
- |
Unigene0005540(GAP1) |
Gene |
- |
- |
- |
|
…… |
…… |
…… |
…… |
…… |
…… |
注:type列pathway表示用户输入的通路,Add_link表示补充出来的连接通路,Add_Adjacent表示补充出来的相邻通路,gene表示用户输入的基因;由于补充通路和基因没有gene number、degree和FDR值,因此用“-”表示。
(2)关系文件:(Edge)
|
fromNode_ID |
fromNode_name |
toNode_ID |
toNode_name |
Relations Type |
|
ko00001 |
|
ko00002 |
|
Path-path |
|
ko00002 |
|
ko00004 |
|
Path-path |
|
ko00005 |
|
ko00006 |
|
Path-path |
|
…… |
…… |
…… |
…… |
…… |
|
ko00007 |
|
ko00008 |
|
Path-link |
|
ko00008 |
|
Ko00006 |
|
Link-path |
|
…… |
…… |
…… |
…… |
…… |
|
ko_00001 |
|
ko_00342 |
|
path-Adjacent |
|
ko_00001 |
|
ko_04521 |
|
path-Adjacent |
|
ko_00001 |
|
ko_12316 |
|
path-Adjacent |
|
ko_00001 |
|
ko_00121 |
|
path-Adjacent |
|
ko_00006 |
|
ko_00543 |
|
path-Adjacent |
|
ko_00006 |
|
ko_00214 |
|
path-Adjacent |
|
ko_00007 |
|
ko_00953 |
|
path-Adjacent |
|
…… |
…… |
…… |
…… |
…… |
|
ko_00001 |
|
- |
Unigene0003256(Rasa4) |
Path-gene |
|
ko_00001 |
|
- |
Unigene0003714(B0563.7) |
Path-gene |
|
ko_00001 |
|
- |
Unigene0005540(GAP1) |
Path-gene |
|
…… |
…… |
…… |
…… |
…… |
注:path-path表示用户输入的通路关系对;Path-link和Link-path表示补充出来的连接通路关系对;path-Adjacent表示所有相邻的补充通路关系对;path-gene表示通路与基因的关系。
(3)图形:
基础网络图:
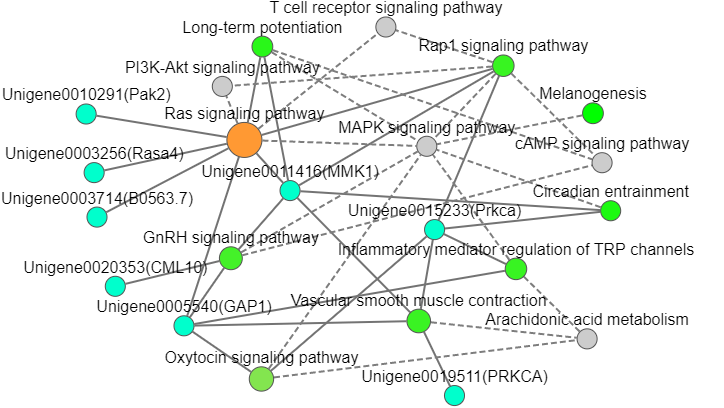
图形解读及说明:
绿色八边形的节点表示基因,它的大小可以根据参数调整单一大小,没有渐变大小;颜色也可以调整单一颜色,没有渐变色。
其他不同颜色和大小的节点表示KEGG pathway,节点大小表示KEGG富集分析后富集到该通路的基因数量,节点的渐变颜色表示KEGG富集分析的Pvalue。
实线表示通路与通路或通路与基因之间有连接关系(说明:KEGG通路没有上下游的关系,只能说他们相互之间存在关系)。
图中孤立的节点表示各通路之间没有关系。
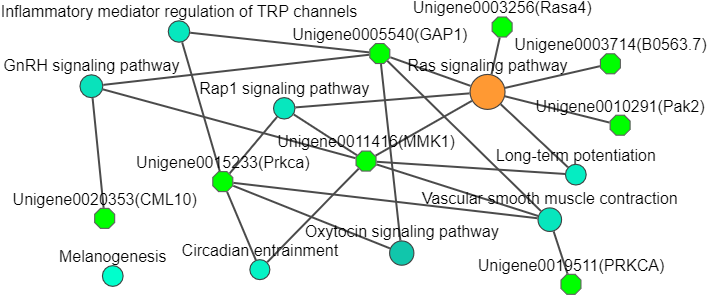
补充连接通路的网络图:

图形解读及说明:
天蓝色圆形的节点表示基因,它的大小可以根据参数调整单一大小,没有渐变大小;颜色也可以调整单一颜色,没有渐变色。
其他不同颜色和大小的节点表示KEGG pathway,节点大小表示KEGG富集分析后富集到该通路的基因数量,节点的渐变颜色表示KEGG富集分析的Pvalue,灰色节点表示从KEGG连接数据库中补充的通路,它的作用是为了连接孤立的通路,使KEGG显著富集的通路形成一个完整的网络,使我们从全局解释生物学问题,了解更多通路信息。
实线表示通路与通路或通路与基因之间有连接关系(说明:KEGG通路没有上下游的关系,只能说他们相互之间存在关系),虚线表示连接补充的通路与输入通路的连接线,形成一个关系网络。
补充相邻通路的网络图:
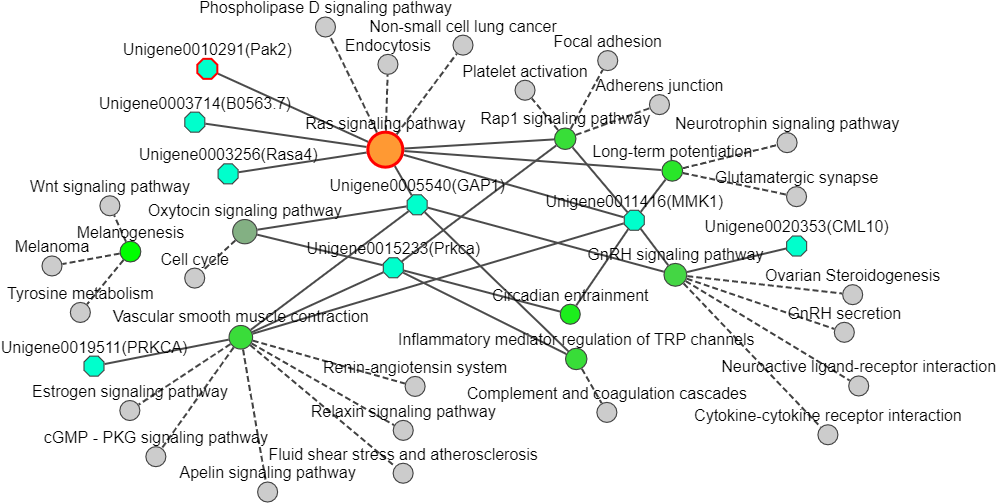
图形解读及说明:
天蓝色八边形的节点表示基因,它的大小可以根据参数调整单一大小,没有渐变大小;颜色也可以调整单一颜色,没有渐变色。红色加粗边框的天蓝色八边形的节点表示该基因在本研究比较重要,用颜色突出显示。
其他不同颜色和大小的节点表示KEGG pathway,节点大小表示KEGG富集分析后富集到该通路的基因数量,节点的渐变颜色表示KEGG富集分析的Pvalue,灰色节点表示从KEGG连接数据库中补充的相邻通路,它可以使我们了解KEGG显著富集的通路的相邻通路。
实线表示通路与通路或通路与基因之间有连接关系(说明:KEGG通路没有上下游的关系,只能说他们相互之间存在关系),虚线表示连接补充的通路与输入通路的连接线,形成一个关系网络。
根据您的经验或查阅文献发现,某通路或基因是与本研究非常相关或很重要,可以通过属性将该节点或连接线重点突出。
补充全部通路可能出现节点和连接线比较多,造成图形整体美观,可以根据自己的需求来做。
Q1. 上传的数据需要保存成什么格式?文件名称和拓展名有没有要求?
OmicShare当前支持txt(制表符分隔)文本文件、csv(逗号分隔)文本文件、以及Excel专用的xlsx格式,同样支持旧版Excel的xls(Excel 97-2003 )格式。如果是核酸、蛋白序列文件,必须为FASTA格式(本质是文本文件)。
文件名可由英文和数字构成,文件拓展名没有限制,可以是“.txt”、“.xlsx”、“.xls”、“.csv”“.fasta”等,例如 mydata01.txt,gene02.xlsx 。
Q2. 提交时报错常见问题:
1.提交时显示X行X列空行/无数据,请先自查表格中是否存在空格或空行,需要删掉。
2.提交时显示列数只有1列,但表格数据不止1列:列间需要用分隔符隔开,先行检查文件是否用了分隔符。
其它提示报错,请先自行根据提示修改;如果仍然无法提交,可通过左侧导航栏的“联系客服”选项咨询OmicShare客服。
Q3. 提交的任务完成后却不出图该怎么办?
主要原因是上传的数据文件存在特殊符号所致。可参考以下建议逐一排查出错原因:
(1)数据中含中文字符,把中文改成英文;
(2) 数据中含特殊符号,例如 %、NA、+、-、()、空格、科学计数、罗马字母等,去掉特殊符号,将空值用数字“0”替换;
(3)检查数据中是否有空列、空行、重复的行、重复的列,特别是行名(一般为gene id)、列名(一般为样本名)出现重复值,如果有删掉。
排查完之后,重新上传数据、提交任务。如果仍然不出图,可通过左侧导航栏的“联系客服”选项咨询OmicShare客服。
Q4.下载的结果文件用什么软件打开?
OmicShare云平台的结果文件(例如,下图为KEGG富集分析的结果文件)包括两种类型:图片文件和文本文件。
图片文件:
为了便于用户对图片进行后期编辑,OmicShare同时提供位图(png)和矢量图(pdf、svg)两种类型的图片。对于矢量图,最常见的是pdf和svg格式,常用Ai(Adobe illustrator)等进行编辑。其中,svg格式的图片可用网页浏览器打开,也可直接在word、ppt中使用。
文本文件:
文本文件的拓展名主要有4种类型:“.os”、“.xls”、“.log”和“.txt”。这些文件本质上都是制表符分隔的文本文件,使用记事本、Notepad++、EditPlus、Excel等文本编辑器直接打开即可。结果文件中,拓展名为“.os”文件为上传的原始数据;“.xls”文件一般为分析生成的数据表格;“.log”文件为任务运行日志文件,便于检查任务出错原因。
Q5. 提交的任务一直在排队怎么办?
提交任务后都需要排队,1分钟后,点击“任务状态刷新”按钮即可。除了可能需运行数天的注释工具,一般工具数十秒即可出结果,如果超出30分钟仍无结果,请联系OS客服,发送任务编号给OmicShare客服,会有专人为你处理任务问题。
Q6. 结果页面窗口有问题,图表加载不出来怎么办?
尝试用谷歌浏览器登录OmicShare查看结果文件,部分浏览器可能不兼容。




 扫码支付更轻松
扫码支付更轻松



