Pfam数据库功能注释简介
蛋白质一般包含1个或多个功能区域,称为结构域。不同结构域的组合产生了丰富的蛋白质,对蛋白质结构域的鉴定也可以推测蛋白质功能。Pfam(Protein families database of alignments and hidden Markov models )提供了完整准确的蛋白质家族和结构域分类信息,以多序列比对信息和隐马尔可夫模型(HMM)表示,广泛应用于蛋白家族查询和蛋白结构域的注释。PfamA 中所包含的蛋白结构数据都是已知并且得到验证的,每个蛋白结构域都有各自的定义(definition)。
使用Pfam_Scan(https://www.ebi.ac.uk/Tools/pfa/pfamscan/)默认参数将目标序列与数据库比对。我们使用的数据库为2018.09更新的最新版Pfam 32.0,包含17,929个蛋白家族信息。
输入文件
上传fasta格式的蛋白序列文件。
1. 比对结果总表
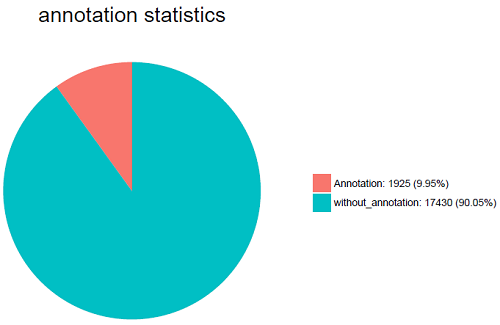
2.比对结果统计饼图将比对上的与比对不上的序列数目进行统计并画饼图。
结果
1. 比对结果总表

表头说明:
Seq id : 蛋白/基因的编号
Alignment start : 基因/蛋白序列比对的结构域起始位置
Alignment end : 基因/蛋白序列比对的结构域终止位置
Envelope start : HMM 模型预测的基因/蛋白序列的结构域起始位置
Envelope end : HMM 模型预测的基因/蛋白序列的结构域终止位置
Hmm acc: 基因/蛋白序列对应结构的模型在Pfam中的编号
Hmm name : 基因/蛋白序列对应结构的模型在Pfam中的名称
Type : 基因/蛋白序列匹配到 Pfam 数据库中对应结构的分类水平,蛋白家族或者结构域
Hmm start : 比对上的部分在数据库匹配序列上的起始位置
Hmm end : 比对上的部分在数据库匹配序列上的终止位置
Hmm length:比对上的长度
Bit score:根据比对和 HMM 模型得出的基因/蛋白序列结构的评分,打分越高,可信度越高
E_value : 比对的 E值(E值越小,可信度越高)
Significance : 基因/蛋白序列在数据库中匹配结构的数目
Clan : Pfam 数据库中按照蛋白质序列,结构以及 HMM 文件而分成的类群
PfamA definition : 查询序列对应结构在 PfamA 中的名称2.比对结果统计饼图
将比对上的与比对不上的序列数目进行统计并画饼图。
Q1. 上传的数据需要保存成什么格式?文件名称和拓展名有没有要求?
OmicShare当前支持txt(制表符分隔)文本文件、csv(逗号分隔)文本文件、以及Excel专用的xlsx格式,同样支持旧版Excel的xls(Excel 97-2003 )格式。如果是核酸、蛋白序列文件,必须为FASTA格式(本质是文本文件)。
文件名可由英文和数字构成,文件拓展名没有限制,可以是“.txt”、“.xlsx”、“.xls”、“.csv”“.fasta”等,例如 mydata01.txt,gene02.xlsx 。
Q2.注释工具的任务一般要跑多久?
注释工具的任务时长一般与提交的序列条数成正比,且不同工具耗时也差异很大。例如GO功能注释近一年来所有任务平均时长约为2天,而NR注释工具近一年来所有任务平均时长约为3小时。一般情况下,如果任务耗时超出一周可联系OS客服,发送任务编号给OmicShare客服,会有专人为你处理任务问题。




 扫码支付更轻松
扫码支付更轻松


