1 全局修改:
2 背景修改:
3 模块编辑:
1.功能:
趋势分析是针对多个连续型样本(至少3个)的特点(样本间包含特定的时间、空间或处理剂量大小顺序)而对基因的表达模式(在多阶段中表达曲线的形状)进行聚类的方法,通过趋势分析可以找到具有相同表达模式的基因以及蛋白或代谢物等。
2.应用范围:
寻找及可视化基因在连续变化的样本或组中丰度的变化趋势,也可以用于蛋白质、代谢物等具有丰度变化的分析。推荐用于3-5组研究。
3. 输入:
(1)输入文件:
输入的表格文件,支持txt(制表符分隔)文本文件、csv(逗号分隔)文本文件、以及Excel专用的xlsx格式,同样支持旧版Excel的xls(Excel 97-2003 )格式。输入源表格文件:基因表达量表,第一列为基因id,后面为样品表达量,趋势顺序按样品在表格中的顺序排列。基因注释信息文件的第一列为基因名,其他列为相应基因的描述信息。
注意:若文件超过5Mb大小则需要在“我的数据”版块中上传本地数据,之后在工具页面选择云端文件进行分析。
(2)趋势数量:
当两个时间点间变化的趋势数量取1的时候,趋势总个数为3^(n-1)-1,n表示样品个数,如3个样品理论上有8个趋势,当点较多时,可以输入控制输出趋势个数,如5个点理论有80个趋势,可以选择输出20个趋势。
(3)数据预处理方法:
a.Log2标准化:输入为基因表达量,与第一个点比较求出log2(FC),即(log2(v1/v1),log2(v2/v1),log2(v3/v1)…),做趋势分析。
b.标准化:直接用数据做趋势分析,即(v1-v1,v2-v1,v3-v1…),做趋势分析。
c.不做标准化/加0:STEM程序会过滤掉没有趋势的情况(即变化一致的情况会被过滤掉),在数据前加0处理,即(0,v1,v2,v3…),就能够输出这种情况(如:平行处理组相对于对照组都是增加了两倍)。
(4)显著趋势P值阈值:
默认P值<0.05的趋势是显著趋势
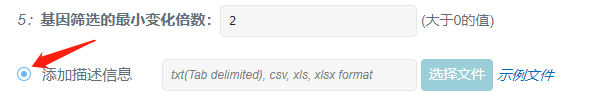
(5)基因筛选的最小变化倍数:
两个点间的表达量低于最小变化趋势会认为没有变化,默认为2倍。
(6)描述信息:
第一列是基因名,其他列是对应的基因注释信息。
4. 结果输出:
程序根据输入的文件进行计算、绘图,过程中等待时间与输入文件大小有关。可以在结果展示中切换任务进行查看。
趋势总览图可以在结果处进行调整。单个模块的结果及阴影图可以在“我的任务”,下载相应任务编号的结果。
输出文件包括:
(1)all_profile.xls:趋势汇总文件
(2)all.xls:所有基因文件
(3)trend_all_by_gene_number.png:显示基因数量的趋势总图
(4)trend_all_by_pvalue.png:显示P值的趋势总图
(5)Profile*.png:单个趋势的图
(6)Profile*.xls:单个趋势的结果文件
文件说明:
(1) trend_all_by_gene_number.png,trend_all_by_pvalue.png 趋势分析结果总览,按照基因数量和p值排序,统计各个趋势的结果。默认是按p值排序的统计结果。
(2) all.xls 基因表达量总表,包含基因id,和各个分组的表达量信息,运行添加注释信息则会包含注释信息。
(3) all_profile.xls 基因趋势总表,各个分组的值为处理后的值,profile表示趋势的序号,SPOT为程序运行时的基因序号。运行添加注释信息则会包含注释信息。
(4) profile[0-n].png,profile[0-n].xls 各个趋势的结果图,以及各个趋势的结果表格(基因id以及各组表达量信息,运行添加注释信息则会包含注释信息).
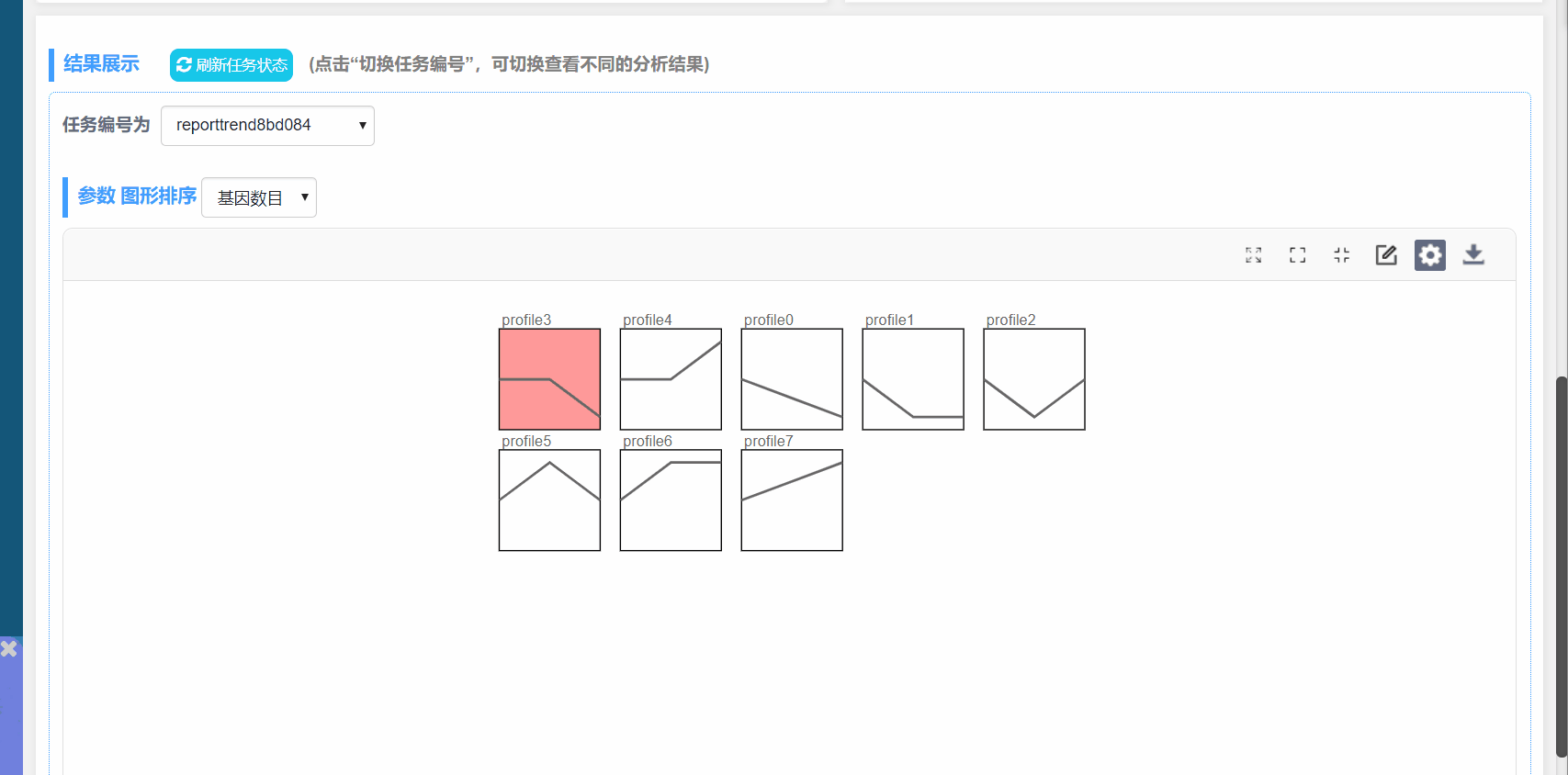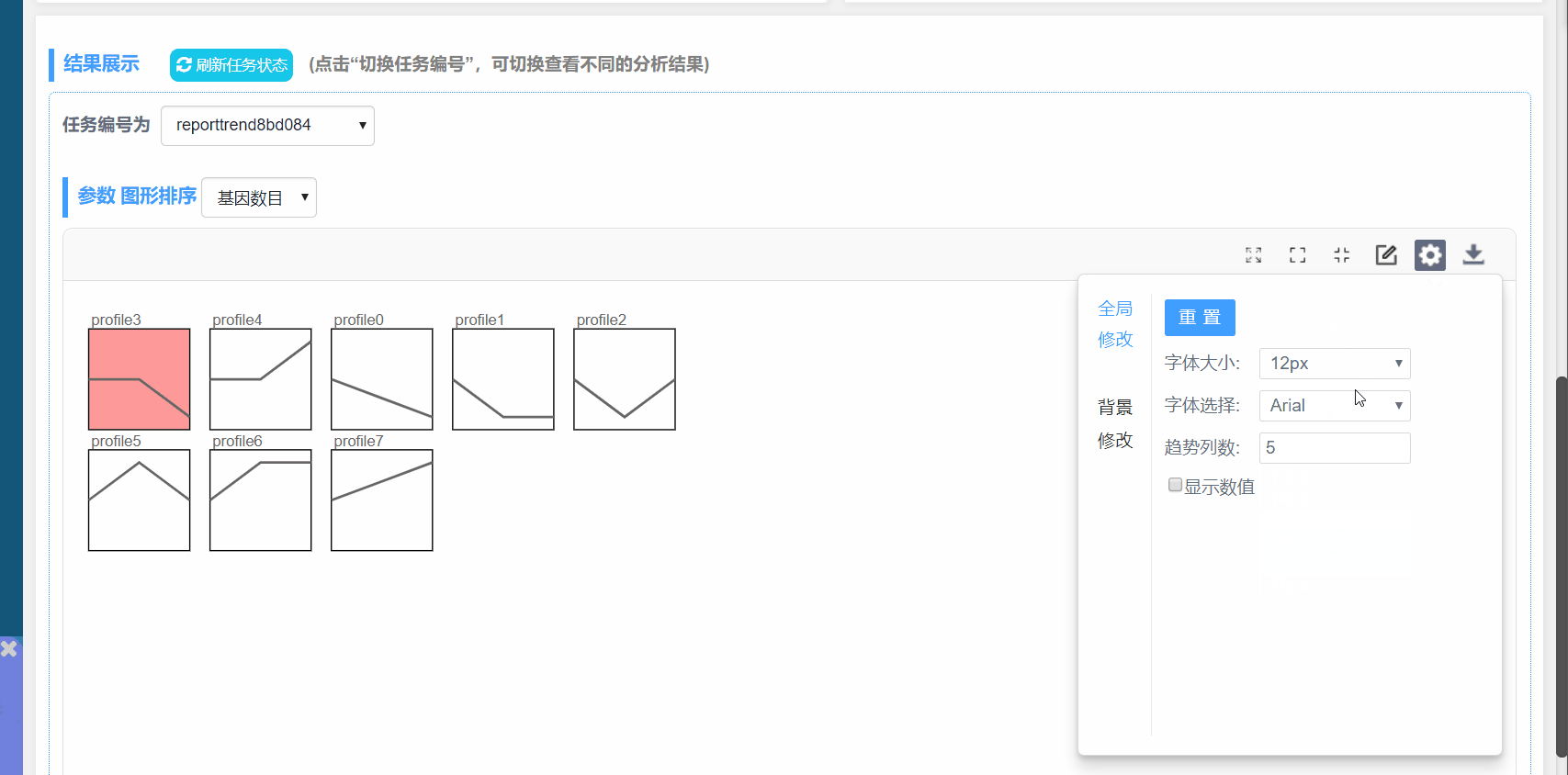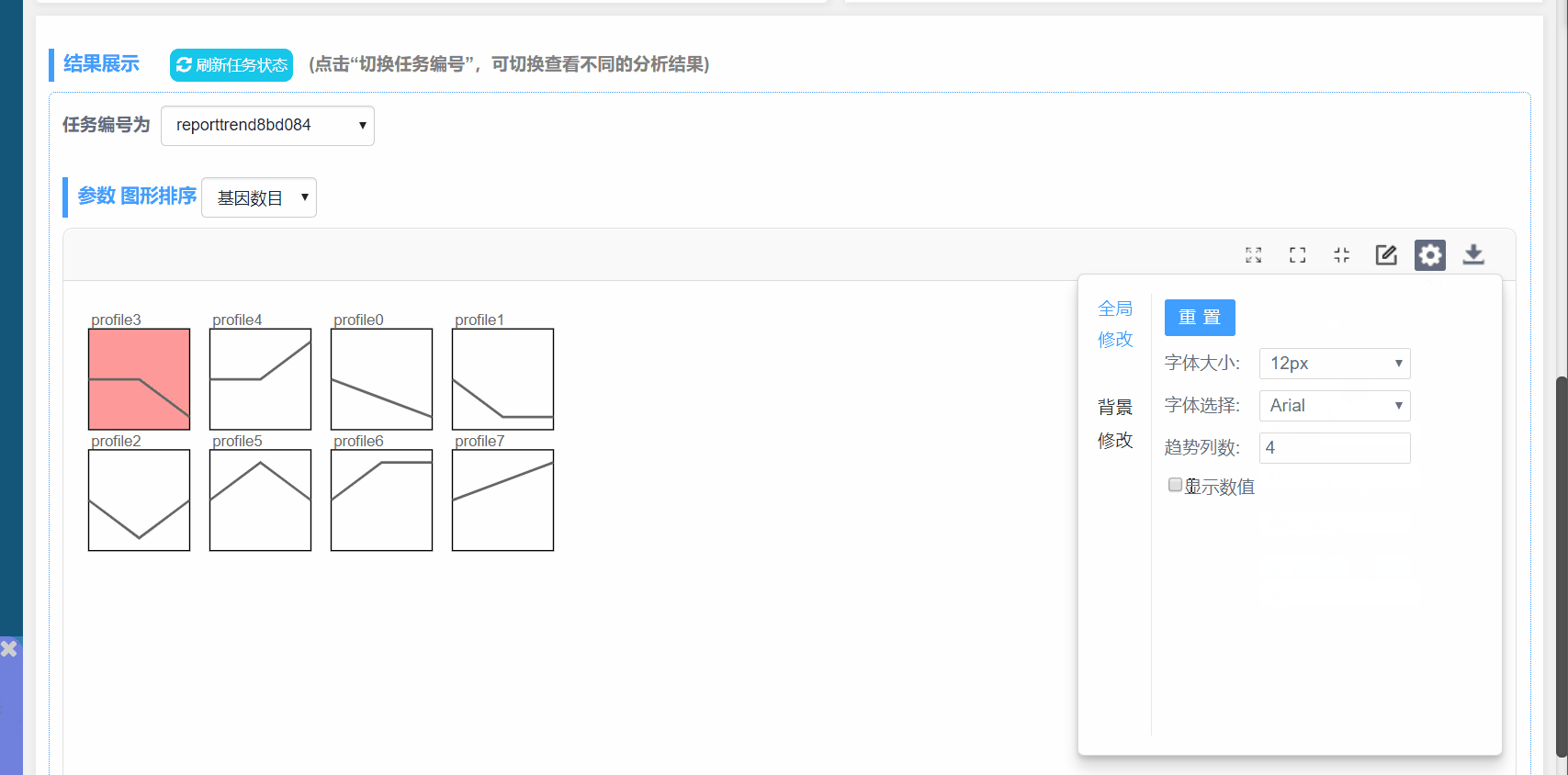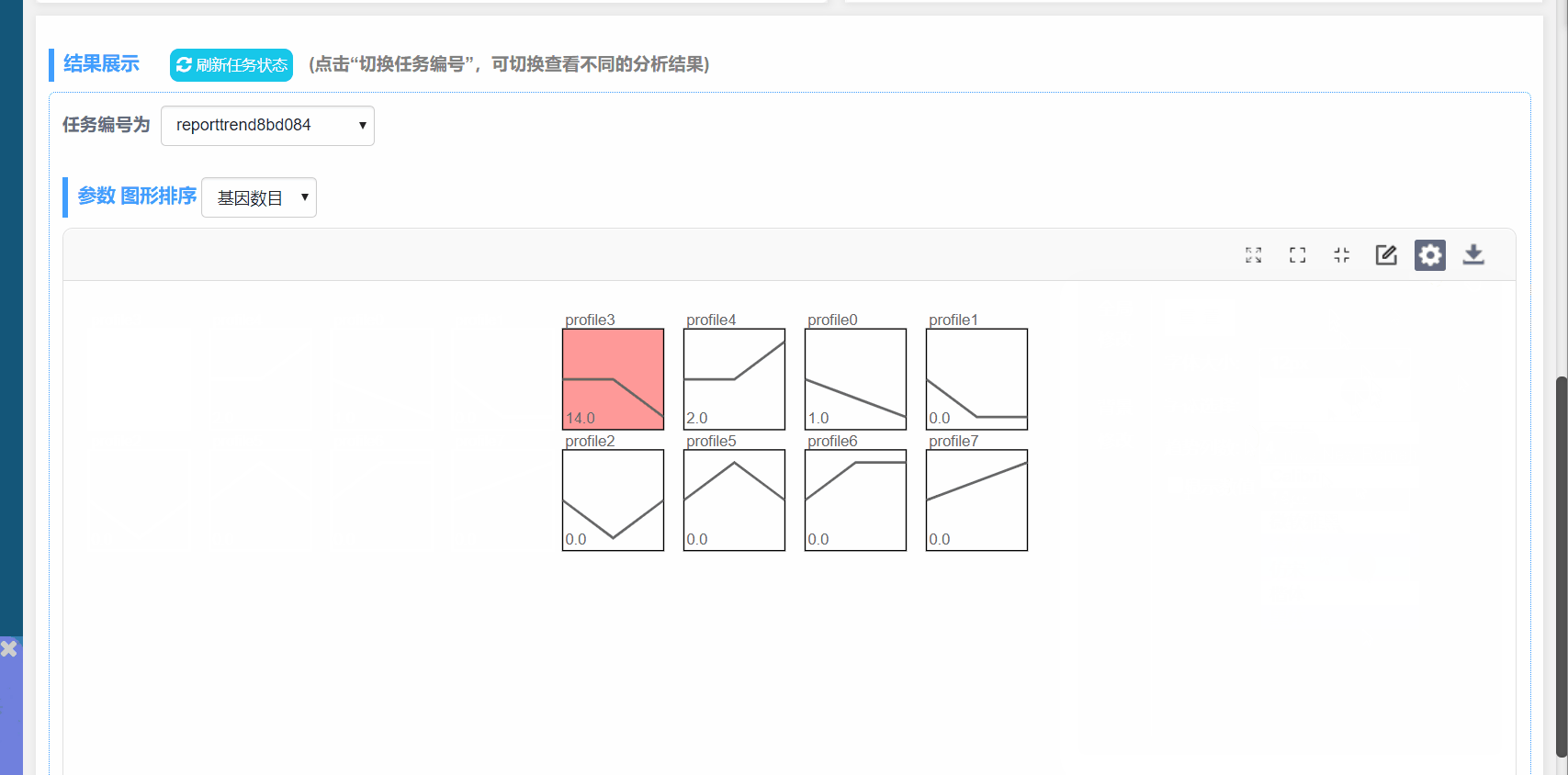
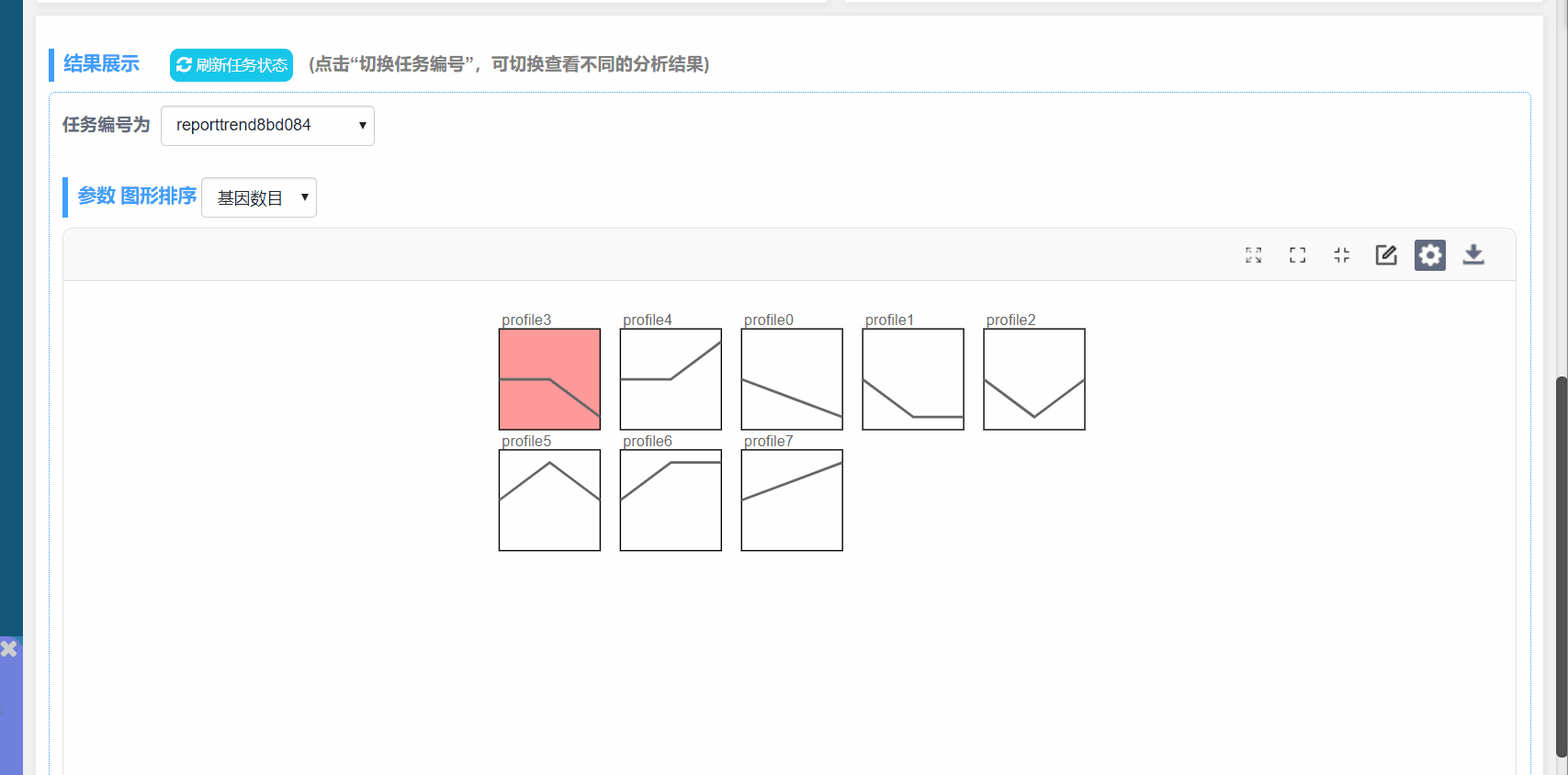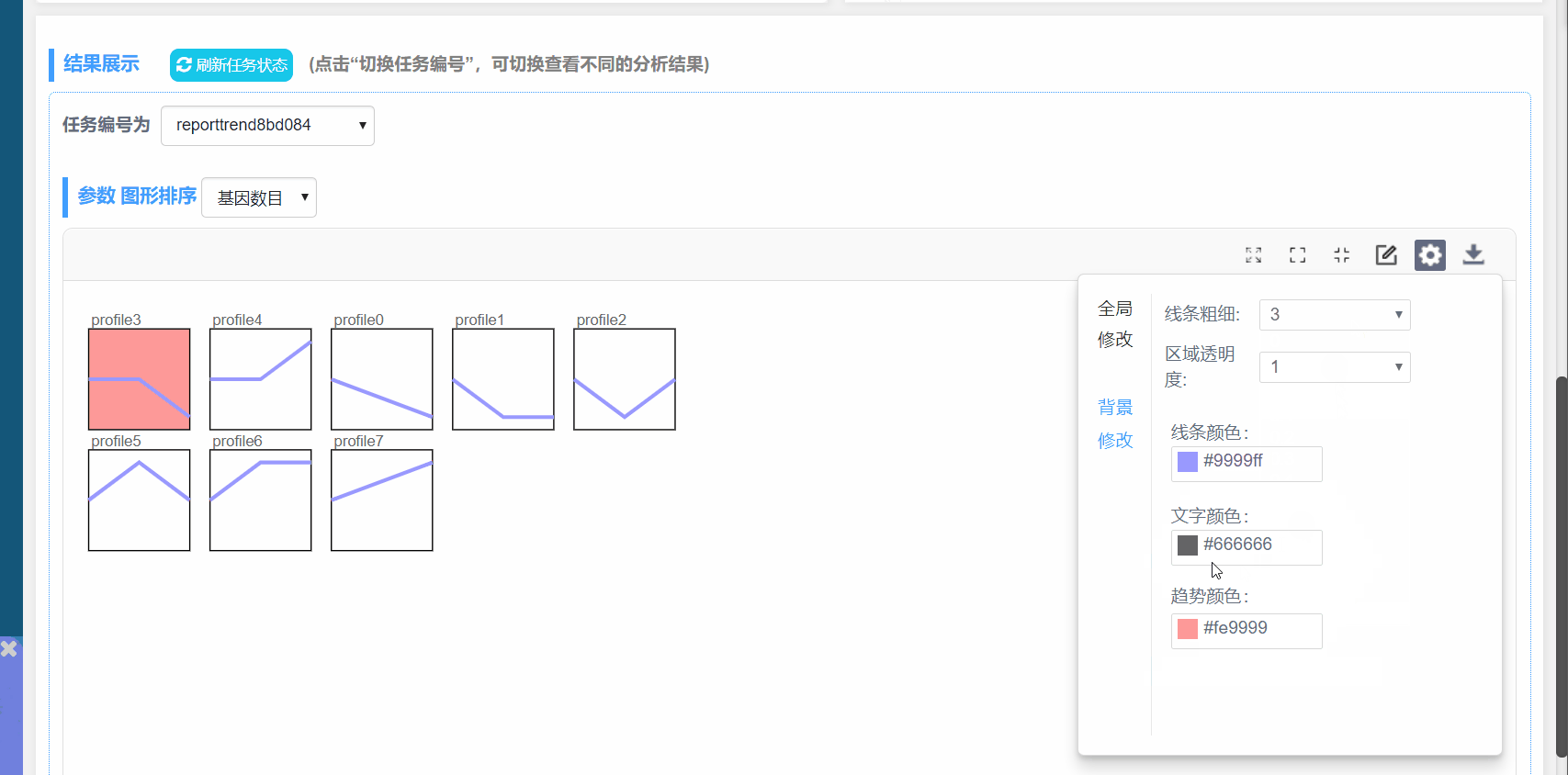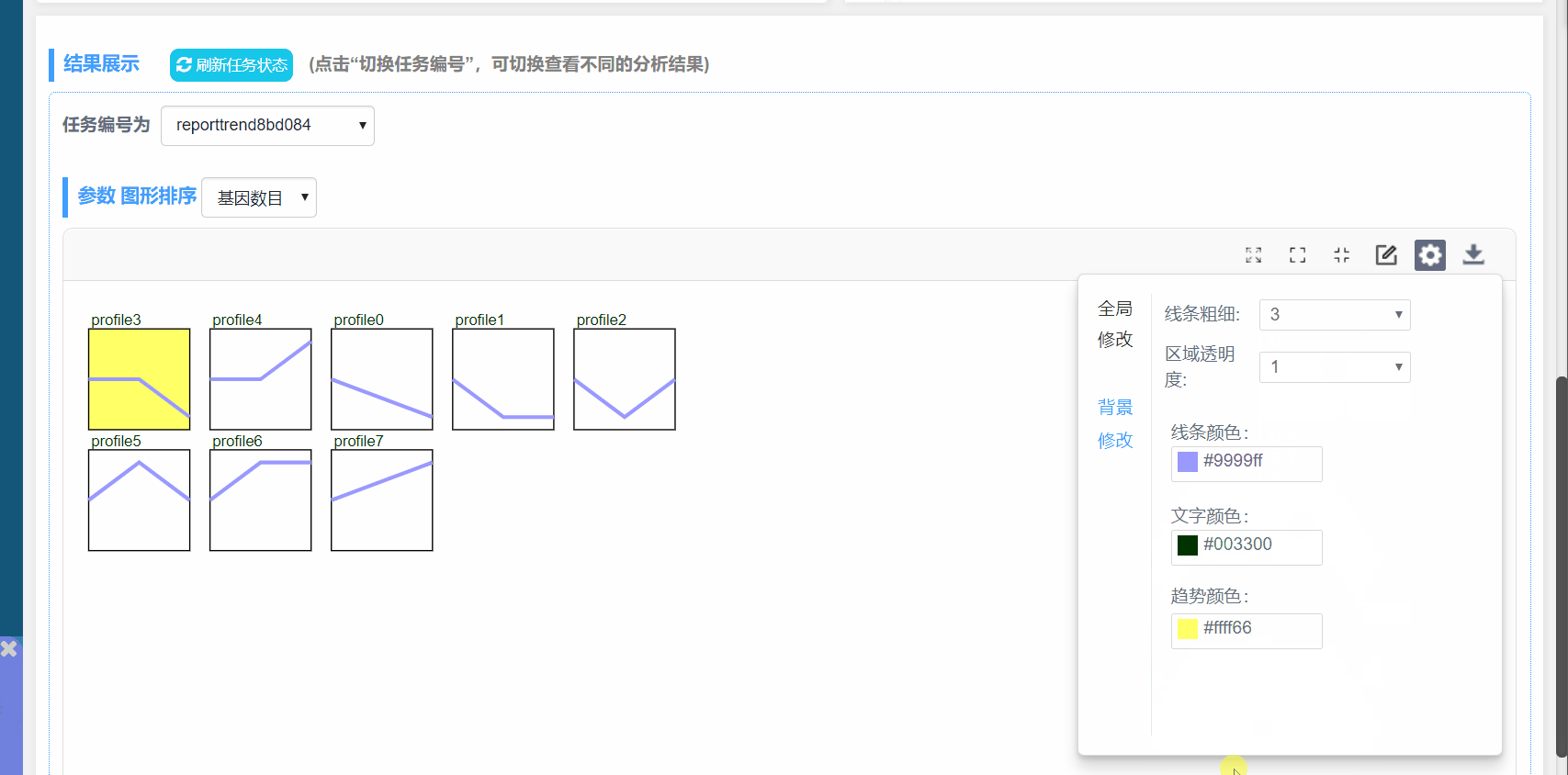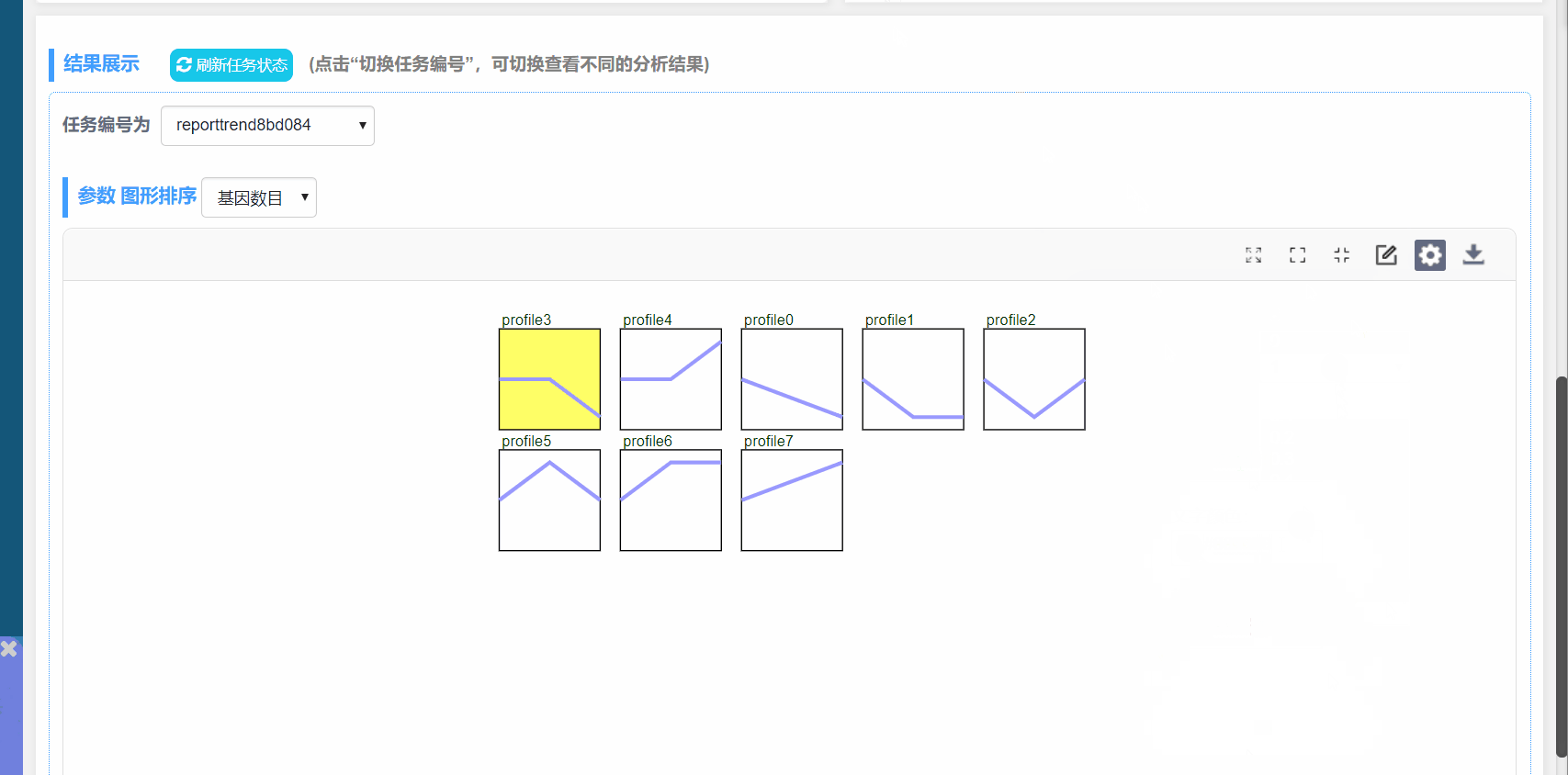
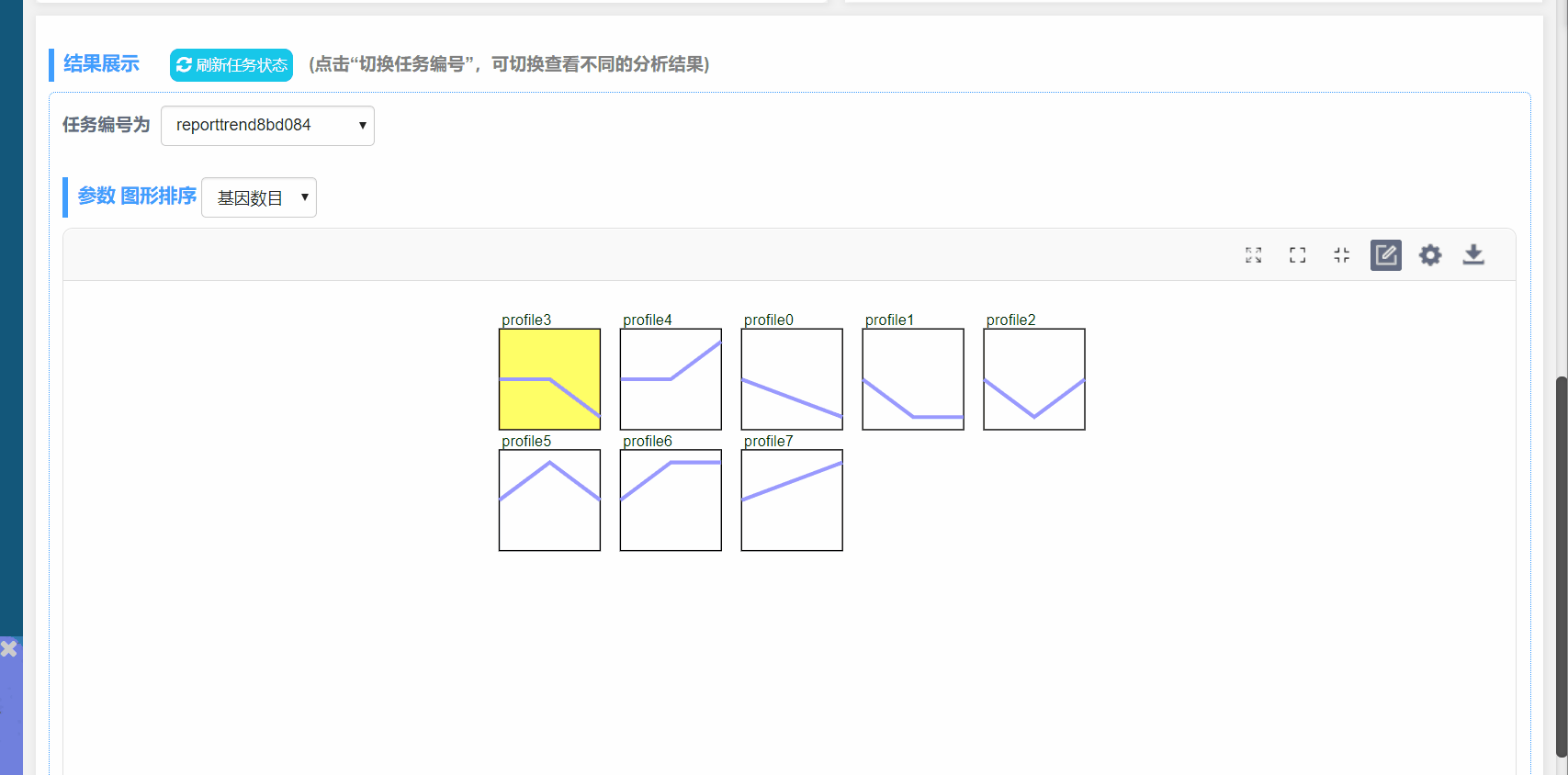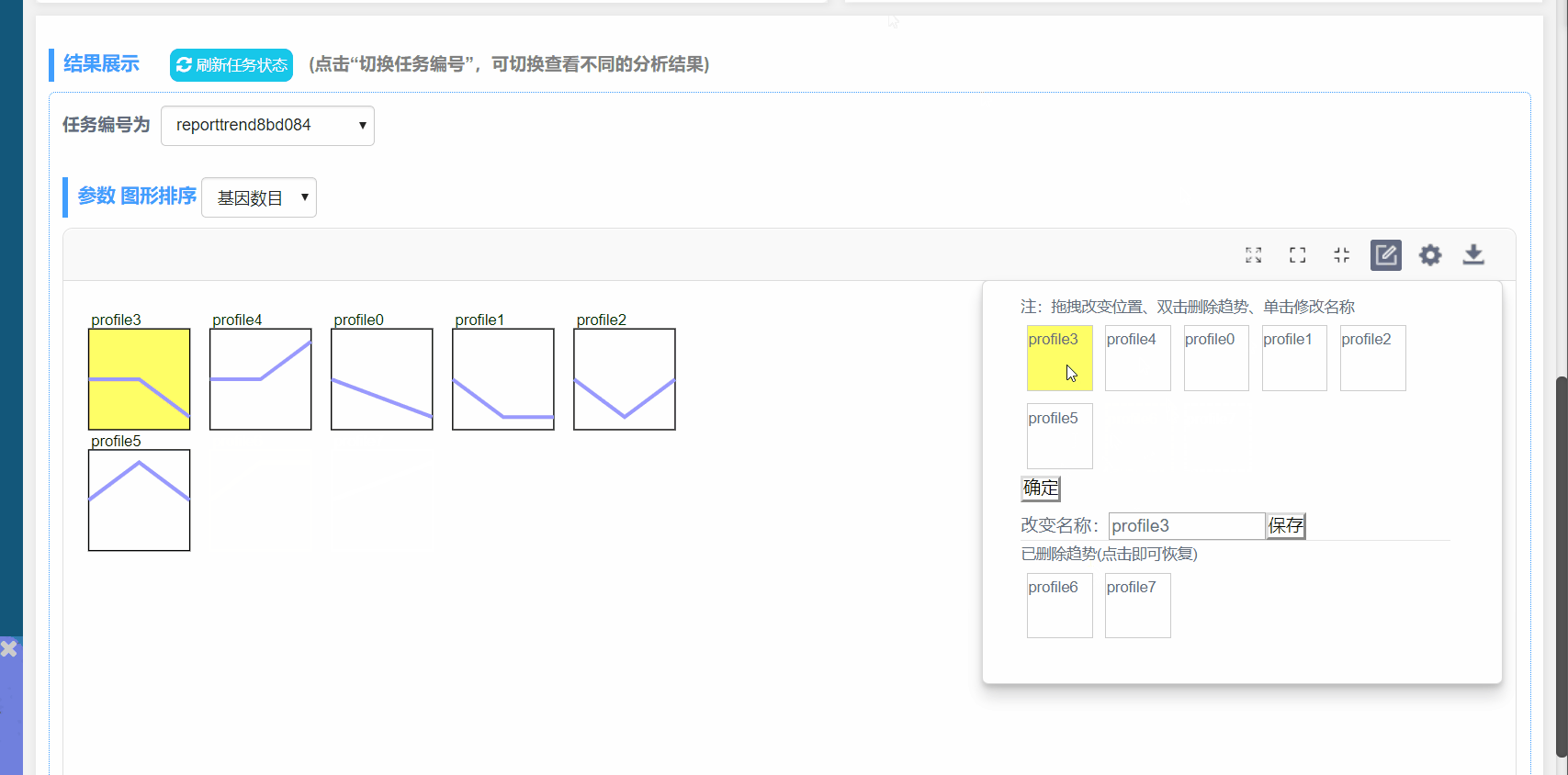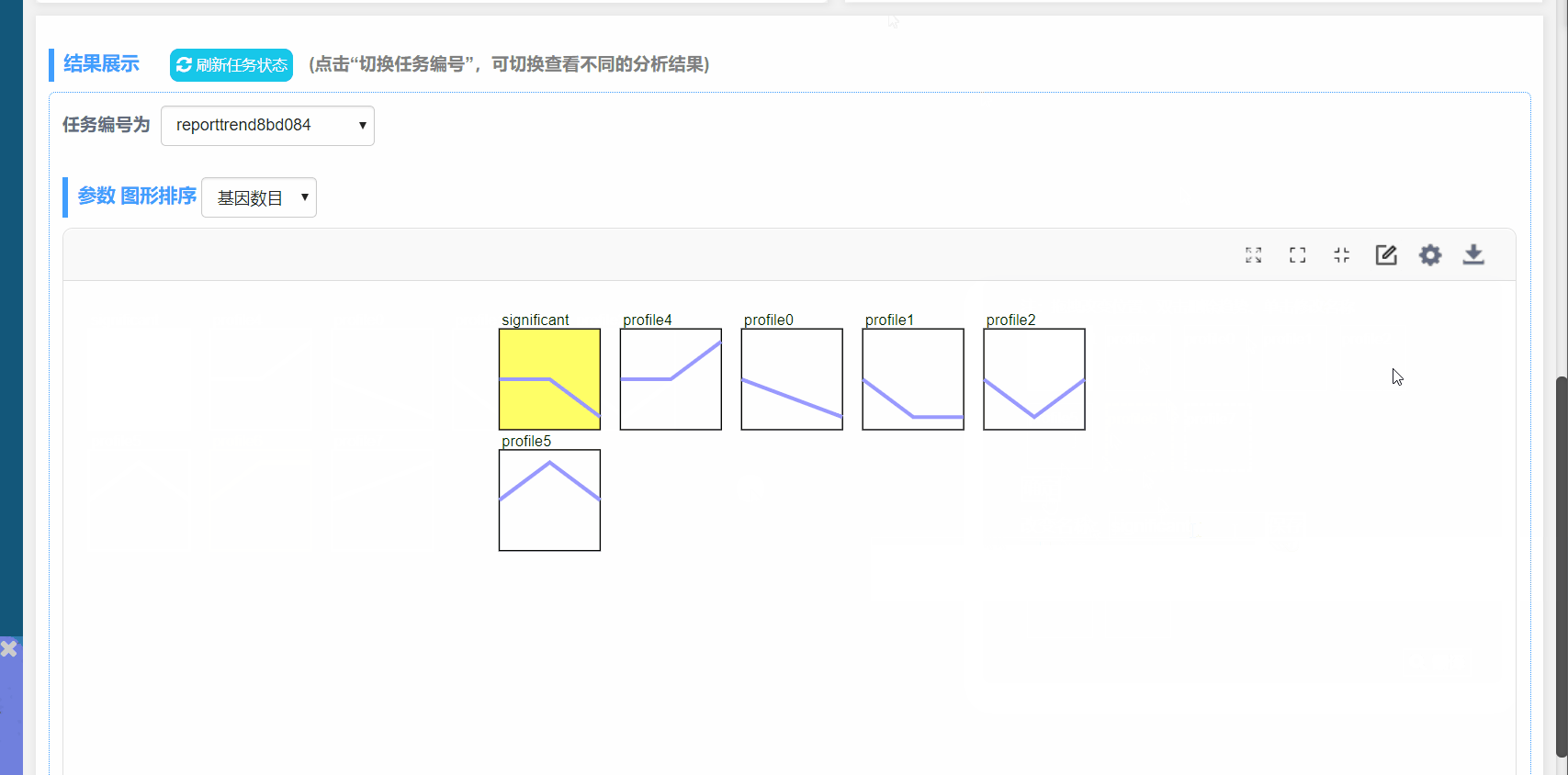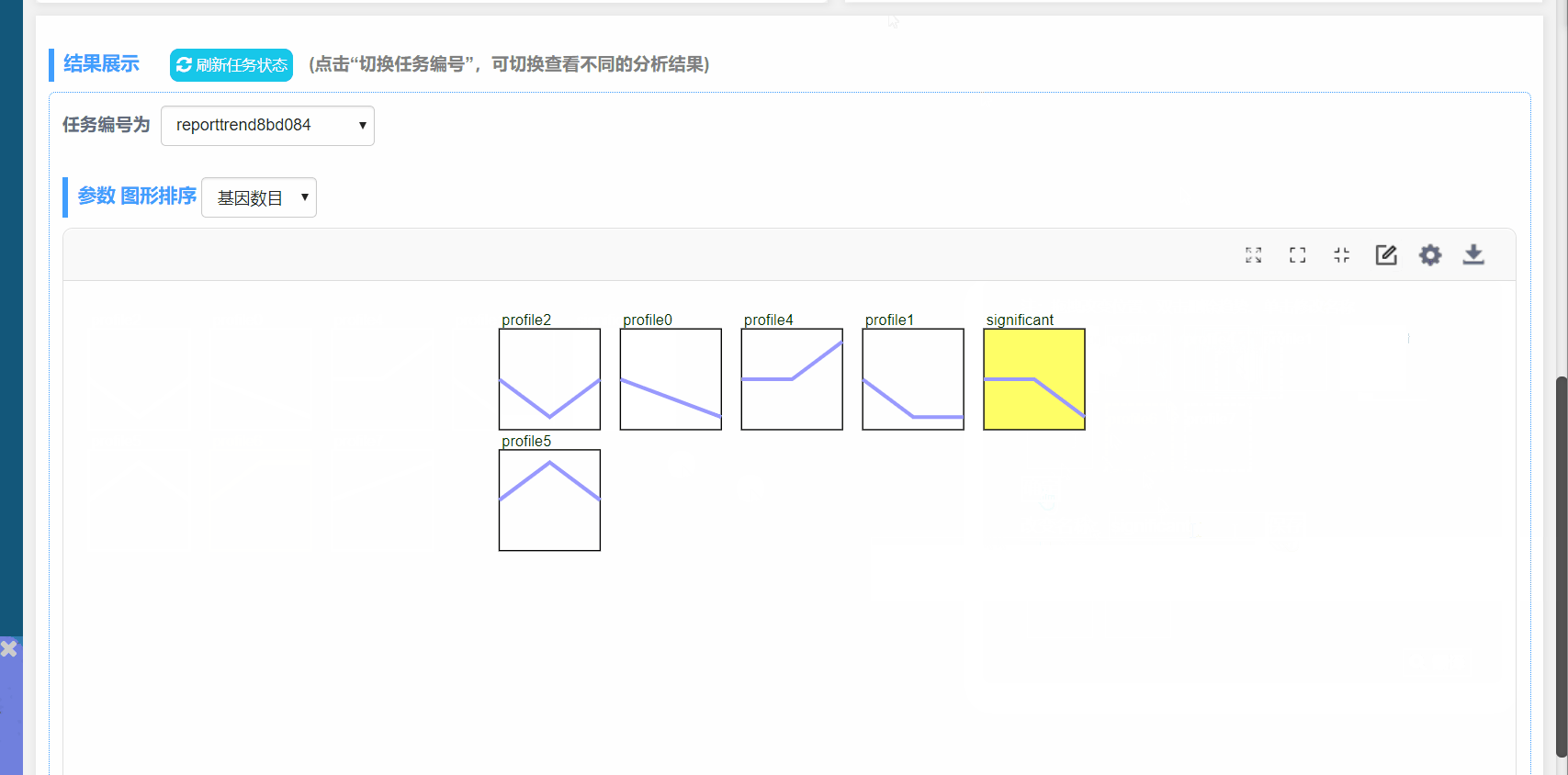
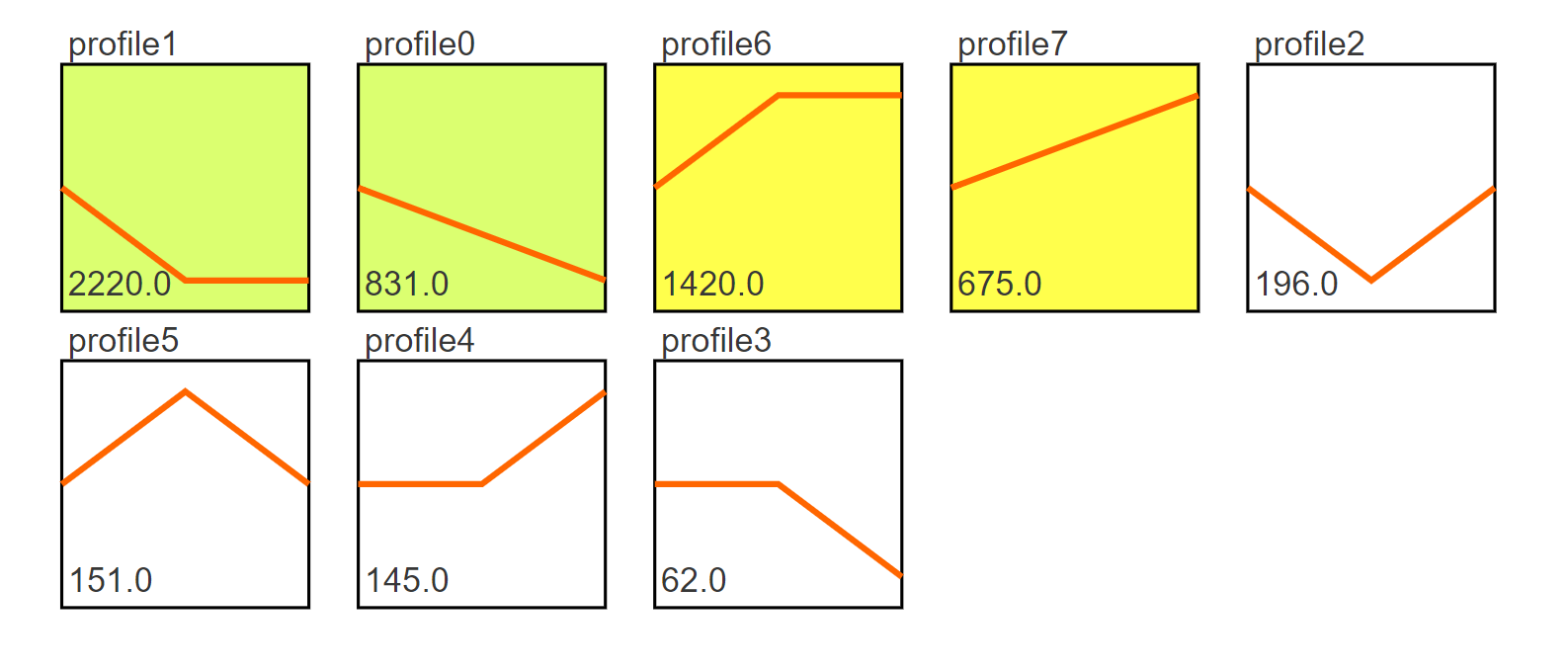
图形解释:
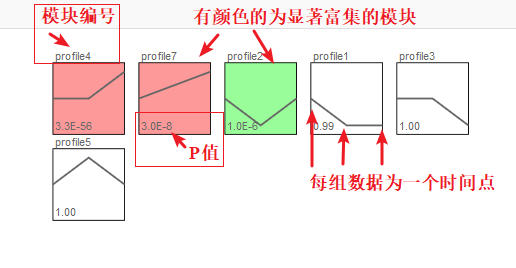
(1) profile n:模块编号
(2) 趋势线:趋势线为每个模块内基因拟合的曲线,每一组为一个拐点
(3) 模块左下角数值:P值,代表富集的显著性
(4) 模块颜色:有颜色模块为p<0.05的模块的显著富集的模块。趋势类似,模块颜色相同,进行分析时可将模块内基因归为一类进行分析,如下图中的profile4和7,都为上升的趋势
5. 图形调整参数说明:
图形调整参数:
字体大小:修改图形内所有字体大小
字体选择:修改图形内所有字体样式
趋势列数:设置一行模块数量
显示数值:勾选显示P值大小
图形排序:根据模块P值或模块内基因数量进行排序
背景修改:
线条粗细:修改模块内线条粗细
区域透明度:修改所有模块背景透明度
线条颜色:修改线条颜色
文字颜色:修改图形中所有文字颜色
趋势颜色:对显著富集的已有颜色的模块,修改背景色
编辑趋势:
(1)双击删除模块,单击可还原
(2)自定义模块顺序
(3)自定义模块名称
下载:
调整图形高度和宽度后,选择“svg"或"png"格式下载结果图。
Q1. 提交时报错常见问题:
1.提交时显示X行X列空行/无数据,请先自查表格中是否存在空格或空行,需要删掉。
2.提交时显示列数只有1列,但表格数据不止1列:列间需要用分隔符隔开,先行检查文件是否用了分隔符。
其它提示报错,请先自行根据提示修改;如果仍然无法提交,可通过左侧导航栏的“联系客服”选项咨询OmicShare客服。
Q2. 趋势分析任务出错?
1.只输入两组样本,运行会出错。趋势分析的目的是看基因表达量变化趋势,三组及以上才能看出波动变化(如持续下调、先上调再下调等),因此必须输入三组及以上连续型样本。
2.使用代谢物等进行分析,名称可能会有大量特殊符号,导致任务出错。
建议:在表达量表中用代号符号(如a1、a2....替换掉名称),然后在“添加描述信息”中,另外上传含代号、代谢物名称的文件,名称等信息会自动填充到结果文件中。
Q3. 提交的任务完成后却不出图该怎么办?
主要原因是上传的数据文件存在特殊符号所致。可参考以下建议逐一排查出错原因:
(1)数据中含中文字符,把中文改成英文;
(2) 数据中含特殊符号,例如 %、NA、+、-、()、空格、科学计数、罗马字母等,去掉特殊符号,将空值用数字“0”替换;
(3)检查数据中是否有空列、空行、重复的行、重复的列,特别是行名(一般为gene id)、列名(一般为样本名)出现重复值,如果有删掉。
排查完之后,重新上传数据、提交任务。如果仍然不出图,可通过左侧导航栏的“联系客服”选项咨询OmicShare客服。
Q4.下载的结果文件用什么软件打开?
OmicShare云平台的结果文件(例如,下图为KEGG富集分析的结果文件)包括两种类型:图片文件和文本文件。
图片文件:
为了便于用户对图片进行后期编辑,OmicShare同时提供位图(png)和矢量图(pdf、svg)两种类型的图片。对于矢量图,最常见的是pdf和svg格式,常用Ai(Adobe illustrator)等进行编辑。其中,svg格式的图片可用网页浏览器打开,也可直接在word、ppt中使用。
文本文件:
文本文件的拓展名主要有4种类型:“.os”、“.xls”、“.log”和“.txt”。这些文件本质上都是制表符分隔的文本文件,使用记事本、Notepad++、EditPlus、Excel等文本编辑器直接打开即可。结果文件中,拓展名为“.os”文件为上传的原始数据;“.xls”文件一般为分析生成的数据表格;“.log”文件为任务运行日志文件,便于检查任务出错原因。
Q5. 提交的任务一直在排队怎么办?
提交任务后都需要排队,1分钟后,点击“任务状态刷新”按钮即可。除了可能需运行数天的注释工具,一般工具数十秒即可出结果,如果超出30分钟仍无结果,请联系OS客服,发送任务编号给OmicShare客服,会有专人为你处理任务问题。
Q6. 结果页面窗口有问题,图表加载不出来怎么办?
尝试用谷歌浏览器登录OmicShare查看结果文件,部分浏览器可能不兼容。
案例1:
发表期刊:Tree physiology
影响因子:4.0
发表时间:2021
Figure 4 . Trend analysis and lncRNA mRNA regulatory networks. ( Schematic diagram of gene expression trends between samples . The trend ID and the number of genes it contains are at the top. T he colored trend blocks indicate a trend of significant enrichment, and different colors indicate different trends. The partial correlation
引用方式:The hierarchical clustering analys es of all differentially expressed lncRNAs and protein coding genes were implemented by the OmicShare tools ( www.omicshare.com/tools ).
参考文献:
Li L, Liu J, Liang Q, et al. Genome-wide analysis of long noncoding RNAs affecting floral bud dormancy in pears in response to cold stress[J]. Tree physiology, 2021, 41(5): 771-790.
案例2:
发表期刊:Frontiers in Oncology
影响因子:4.7
发表时间:2022
FIGURE 1 Dynamic trend analysis: (A) the significance and gene number of each module; (B) histogram showing the situation of each module.
引用方式:Trend analysis can classify genes with similar change characteristic patterns within a changing trend. We used the OmicShare online tool (https://www.omicshare.com/tools/) for analysis. P <.05 was considered statistically significant. The number of trends is chosen to be 20.
参考文献:
Gao W, Yang M. Identification by Bioinformatics Analysis of Potential Key Genes Related to the Progression and Prognosis of Gastric Cancer[J]. Frontiers in Oncology, 2022, 12: 881015.
案例3:
发表期刊:BMC Plant Biology
影响因子:5.3
发表时间:2023
Fig 8.Trend analysis of DEGs enrichment under different nitrogen levels. (a,b) Venn diagram analysis. (c,d) Trend analysis of differentially expressed genes. The 15,023 DEGs from NL-Al, NL-WT and NH-Al were clustered into 8 profiles. The 11,483 DEGs from NL-SG, NL-WT and NH-SG were clustered into 8 profiles. (e,f) Top 30 of KEGG pathway enrichment of DEGs in Profile 6
引用方式:
Venn diagrams and heatmaps were generated using OmicShare tools, a free online platform for data analysis (https://www.omicshare.com/tools). All DEGs were mapped to GO terms in the Gene Ontology database [48]. The KEGG pathways were used to assign DEGs with the online KEGG automatic annotation server (KAAS) [49,50,51]. An FDR below 0.05 was the threshold to identify significant GO terms and enriched pathways in DEGs. Then, the KEGG network was also generated using OmicShare tools.
To further explore the regulatory network of gene expression associated with the phenotype of seedlings, gene co-expression networks of 24,882 genes of which FPKM values > 5.8 were constructed using OmicShare tools.
参考文献:
Feng Y, Li Y, Zhao Y, et al. Physiological, transcriptome and co-expression network analysis of chlorophyll-deficient mutants in flue-cured tobacco[J]. BMC Plant Biology, 2023, 23(1): 1-16.
案例4:
发表期刊:Frontiers in Plant Science
影响因子:5.6
发表时间:2021
FIGURE 2. Trend analysis of the differentially expressed target genes of specific miRNAs in (A) roots and (B) leaves, and profiles are ordered based on the number of genes assigned.
引用方式:The trend analysis was performed using the OmicShare tools.
参考文献:
Nie G, Liao Z, Zhong M, et al. MicroRNA-mediated responses to chromium stress provide insight into tolerance characteristics of Miscanthus sinensis[J]. Frontiers in Plant Science, 2021, 12: 666117.
案例5:
发表期刊:Frontiers in Plant Science
影响因子:5.6
发表时间:2021
Figure 3 Trend analysis of colorectal cancer development at different stages of cancer progression. Each row in the figure represents the clustering results of each tissue type (seven profiles were obtained); from top to bottom: tumor tissues, paracancerous tissues, and normal tissues. The lower left corner number represents the number of metabolites with the trend, and the upper right corner represents the significant results of these metabolites clustering into the trend (colored trend indicates that the difference was statistically significant).
引用方式:Short Time-series Expression Mine software (STEM) (performed on OmicShare tools platform; www.omicshare.com/tools) simulates the possible change trends and then calculates the correlation coefficient between each factor and the preset trend, and divides the factors into the most similar trends. We preprocessed the data by log2 and set 1.2 as the minimum fold change of the trend. Finally, a Venn diagram was analyzed.
参考文献:
Lin L, Zeng X, Liang S, et al. Biomarkers of coordinate metabolic reprogramming and the construction of a co-expression network in colorectal cancer[J]. Annals of Translational Medicine, 2022, 10(20).
案例6:
发表期刊:Frontiers in Genetics
影响因子:3.7
发表时间:2022
FIGURE 2. Analysis of gene expression trend in fruit tissue at six developmental stages. (A). Profiles are ordered based on the p-value significance of gene assigned. (B). The level of the gene expression at six developmental stages in profile 0. (C). The level of gene expression at six developmental stages in profile 79.
引用方式:
The gene expression levels in each sample were converted into a data matrix, and PCA was analyzed using the OmicShare tools, a free online platform for data analysis (https://www.omicshare.com/tools). A p-value of 0.05 and the absolute value of |log2 (Fold Change)| ≥ 1 were set as the threshold for significantly differential expression (Audic and Claverie, 1997).
To investigate patterned differences in expression profiles during fruit developmental stages, we analyzed dynamic expression trends of genes using the OmicShare tools. Gene expression was normalized with log2(FC) and gene expression trends were set to 80 profiles using the Dynamic Trend Analysis tool with a p-value of significant expression trend < 0.05 and change multiple > 2. We extracted the expression of all genes in the significant profiles.
参考文献:
Hu P, Zhao M, Chen S, et al. Transcriptional regulation mechanism of flavonoids biosynthesis gene during fruit development in Astragalus membranaceus[J]. Frontiers in Genetics, 2022, 13: 972990.
其他参考文献:
Wang T, Xing L, Song H, et al. Large-scale metabolome analysis reveals dynamic changes of metabolites during foxtail millet grain filling[J]. Food Research International, 2023, 165: 112516.
Wang T, Xing L, Song H, et al. Large-scale metabolome analysis reveals dynamic changes of metabolites during foxtail millet grain filling[J]. Food Research International, 2023, 165: 112516.
案例7:
发表期刊:Ecotoxicology and Environmental Safety
影响因子:6.8
发表时间:2023
Fig. 2. Series test of cluster (STC) analysis of experiment A and experiment B DEGs. The STC result of experiment A DEGs was listed on the left (A), and the STC result of experiment B DEGs was listed on the right (B). The number on the upper left of the box denoted different trend profiles. The number on the bottom of the box denoted gene numbers enriched in this profile, and different color boxes indicated significant expression patterns (P < 0.05).
参考文献:
Yu H, Song W, Chen X, et al. Transcriptomic analysis reveals up-regulated histone genes may play a key role in zebrafish embryo-larvae response to Bisphenol A (BPA) exposure[J]. Ecotoxicology and Environmental Safety, 2023, 252: 114578.




 扫码支付更轻松
扫码支付更轻松